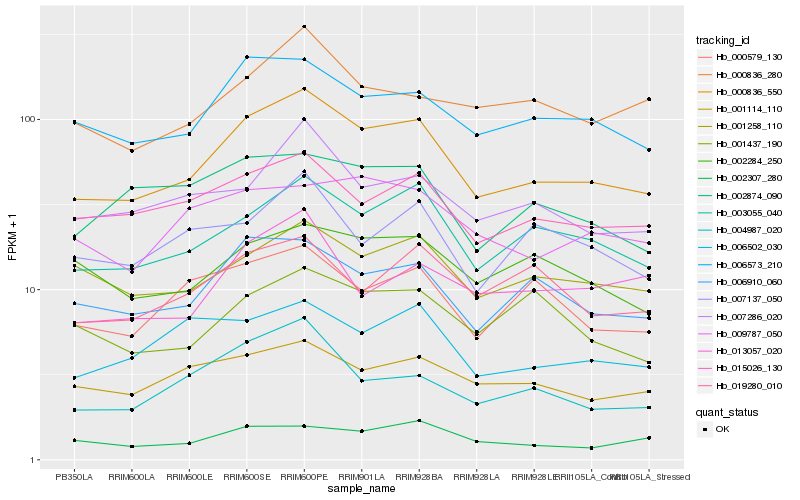
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000836_550 |
0.0 |
- |
- |
PREDICTED: reticulon-like protein B5 [Jatropha curcas] |
| 2 |
Hb_003055_040 |
0.0868914635 |
- |
- |
PREDICTED: secretory carrier-associated membrane protein 3-like isoform X1 [Jatropha curcas] |
| 3 |
Hb_006573_210 |
0.0955581699 |
- |
- |
PREDICTED: protein SGT1 homolog [Jatropha curcas] |
| 4 |
Hb_006910_060 |
0.0977950619 |
- |
- |
PREDICTED: MACPF domain-containing protein At1g14780-like [Jatropha curcas] |
| 5 |
Hb_015026_130 |
0.1050992647 |
- |
- |
quinone oxidoreductase, putative [Ricinus communis] |
| 6 |
Hb_001258_110 |
0.1097550827 |
- |
- |
transporter, putative [Ricinus communis] |
| 7 |
Hb_004987_020 |
0.1118241334 |
- |
- |
- |
| 8 |
Hb_007286_020 |
0.1127662784 |
- |
- |
hypothetical protein L484_010675 [Morus notabilis] |
| 9 |
Hb_000579_130 |
0.1159035203 |
- |
- |
ubiquitin-protein ligase, putative [Ricinus communis] |
| 10 |
Hb_001114_110 |
0.1159427482 |
- |
- |
PREDICTED: probable ubiquitin-like-specific protease 2A isoform X1 [Jatropha curcas] |
| 11 |
Hb_006502_030 |
0.1186191801 |
- |
- |
PREDICTED: probable kinetochore protein ndc80 [Jatropha curcas] |
| 12 |
Hb_002307_280 |
0.1202655919 |
- |
- |
Endonuclease/exonuclease/phosphatase family protein isoform 3 [Theobroma cacao] |
| 13 |
Hb_000836_280 |
0.1215761654 |
- |
- |
PREDICTED: protein BPS1, chloroplastic [Jatropha curcas] |
| 14 |
Hb_019280_010 |
0.1218643937 |
- |
- |
PREDICTED: F-box protein At5g39450 isoform X2 [Jatropha curcas] |
| 15 |
Hb_009787_050 |
0.1227739885 |
- |
- |
(Di)nucleoside polyphosphate hydrolase, putative [Ricinus communis] |
| 16 |
Hb_002874_090 |
0.122913509 |
- |
- |
PREDICTED: uncharacterized protein LOC105637452 [Jatropha curcas] |
| 17 |
Hb_013057_020 |
0.124298503 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RLIM-like [Jatropha curcas] |
| 18 |
Hb_001437_190 |
0.1248112342 |
- |
- |
ABC transporter family protein [Hevea brasiliensis] |
| 19 |
Hb_002284_250 |
0.124820047 |
- |
- |
PREDICTED: epidermal growth factor receptor substrate 15-like 1 [Jatropha curcas] |
| 20 |
Hb_007137_050 |
0.1252839861 |
- |
- |
Uncharacterized protein isoform 1 [Theobroma cacao] |