| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
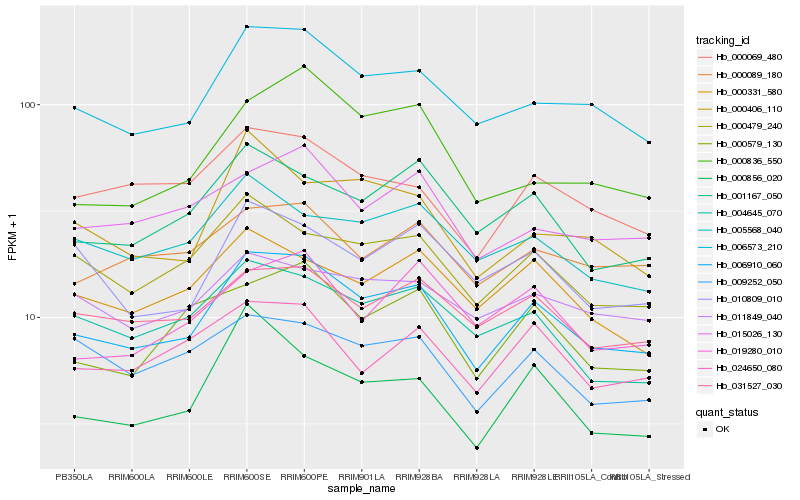
Hb_006910_060 |
0.0 |
- |
- |
PREDICTED: MACPF domain-containing protein At1g14780-like [Jatropha curcas] |
| 2 |
Hb_006573_210 |
0.0600166644 |
- |
- |
PREDICTED: protein SGT1 homolog [Jatropha curcas] |
| 3 |
Hb_000479_240 |
0.0812661987 |
- |
- |
PREDICTED: probable ubiquitin conjugation factor E4 isoform X2 [Jatropha curcas] |
| 4 |
Hb_001167_050 |
0.0841590488 |
- |
- |
PREDICTED: uncharacterized protein LOC105648014 [Jatropha curcas] |
| 5 |
Hb_000579_130 |
0.0853143773 |
- |
- |
ubiquitin-protein ligase, putative [Ricinus communis] |
| 6 |
Hb_031527_030 |
0.086854077 |
- |
- |
PREDICTED: ribosomal RNA processing protein 1 homolog [Jatropha curcas] |
| 7 |
Hb_000069_480 |
0.0872111782 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_004645_070 |
0.0903431898 |
- |
- |
PREDICTED: beta-1,3-galactosyltransferase 15 [Jatropha curcas] |
| 9 |
Hb_024650_080 |
0.0905345014 |
- |
- |
PREDICTED: BTB/POZ domain-containing protein At3g09030 [Jatropha curcas] |
| 10 |
Hb_015026_130 |
0.090584435 |
- |
- |
quinone oxidoreductase, putative [Ricinus communis] |
| 11 |
Hb_000331_580 |
0.0926979901 |
- |
- |
PREDICTED: programmed cell death protein 4-like [Jatropha curcas] |
| 12 |
Hb_005568_040 |
0.0933197686 |
- |
- |
PREDICTED: uncharacterized protein LOC105628163 isoform X2 [Jatropha curcas] |
| 13 |
Hb_010809_010 |
0.0945309648 |
- |
- |
Na+/H+ antiporter [Populus euphratica] |
| 14 |
Hb_000089_180 |
0.0945908759 |
- |
- |
PREDICTED: homoserine kinase-like [Jatropha curcas] |
| 15 |
Hb_000406_110 |
0.0962591384 |
- |
- |
PREDICTED: transmembrane protein adipocyte-associated 1 [Jatropha curcas] |
| 16 |
Hb_011849_040 |
0.0970391069 |
- |
- |
ubiquitin-protein ligase, putative [Ricinus communis] |
| 17 |
Hb_009252_050 |
0.0970772968 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 18 |
Hb_000856_020 |
0.097429354 |
- |
- |
PREDICTED: probable beta-1,3-galactosyltransferase 19 [Jatropha curcas] |
| 19 |
Hb_019280_010 |
0.0974651868 |
- |
- |
PREDICTED: F-box protein At5g39450 isoform X2 [Jatropha curcas] |
| 20 |
Hb_000836_550 |
0.0977950619 |
- |
- |
PREDICTED: reticulon-like protein B5 [Jatropha curcas] |