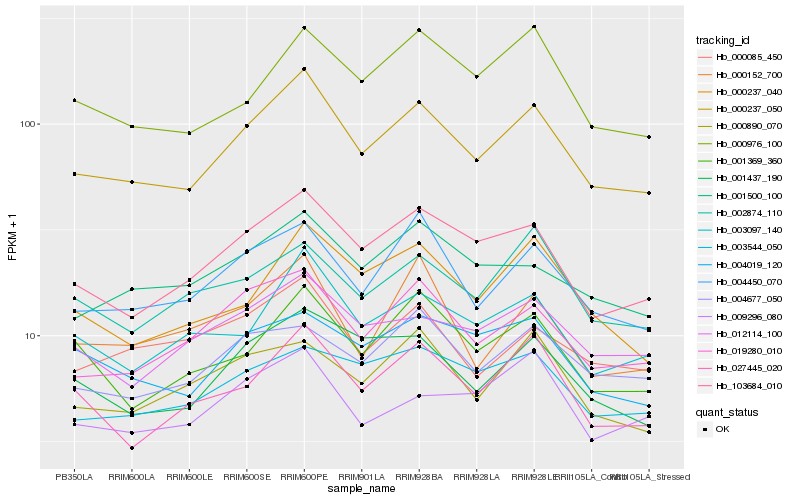
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000237_050 |
0.0 |
- |
- |
CP2 [Hevea brasiliensis] |
| 2 |
Hb_103684_010 |
0.0678757454 |
- |
- |
PREDICTED: cullin-1 isoform X3 [Nicotiana tomentosiformis] |
| 3 |
Hb_004450_070 |
0.0712447423 |
- |
- |
PREDICTED: 2-hydroxyacyl-CoA lyase [Jatropha curcas] |
| 4 |
Hb_019280_010 |
0.0731414261 |
- |
- |
PREDICTED: F-box protein At5g39450 isoform X2 [Jatropha curcas] |
| 5 |
Hb_003097_140 |
0.0762539411 |
- |
- |
PREDICTED: glycosyltransferase-like KOBITO 1 [Jatropha curcas] |
| 6 |
Hb_027445_020 |
0.0770892747 |
- |
- |
PREDICTED: glycosylphosphatidylinositol anchor attachment 1 protein [Jatropha curcas] |
| 7 |
Hb_001437_190 |
0.079127363 |
- |
- |
ABC transporter family protein [Hevea brasiliensis] |
| 8 |
Hb_000237_040 |
0.0813337597 |
- |
- |
PREDICTED: V-type proton ATPase subunit a3-like [Jatropha curcas] |
| 9 |
Hb_000976_100 |
0.0834074937 |
- |
- |
eukaryotic translation elongation factor, putative [Ricinus communis] |
| 10 |
Hb_012114_100 |
0.0836857137 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase At5g41260 [Jatropha curcas] |
| 11 |
Hb_001369_360 |
0.0840887556 |
- |
- |
embryo yellow protein, partial [Manihot esculenta] |
| 12 |
Hb_001500_100 |
0.0863910048 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_000085_450 |
0.0888607401 |
transcription factor |
TF Family: WRKY |
PREDICTED: probable WRKY transcription factor 20 [Jatropha curcas] |
| 14 |
Hb_004677_050 |
0.0905144259 |
- |
- |
PREDICTED: importin beta-like SAD2 isoform X2 [Jatropha curcas] |
| 15 |
Hb_000890_070 |
0.0906018074 |
- |
- |
PREDICTED: protein ALWAYS EARLY 3 isoform X2 [Jatropha curcas] |
| 16 |
Hb_002874_110 |
0.0907983387 |
- |
- |
PREDICTED: uncharacterized protein LOC105637451 [Jatropha curcas] |
| 17 |
Hb_004019_120 |
0.0920714688 |
- |
- |
PREDICTED: conserved oligomeric Golgi complex subunit 1 [Jatropha curcas] |
| 18 |
Hb_003544_050 |
0.0926085131 |
- |
- |
PREDICTED: uncharacterized protein LOC105644300 isoform X1 [Jatropha curcas] |
| 19 |
Hb_009296_080 |
0.0932675398 |
- |
- |
PREDICTED: probable apyrase 7 isoform X1 [Jatropha curcas] |
| 20 |
Hb_000152_700 |
0.0936974144 |
- |
- |
PREDICTED: E3 ubiquitin protein ligase RIN2 [Jatropha curcas] |