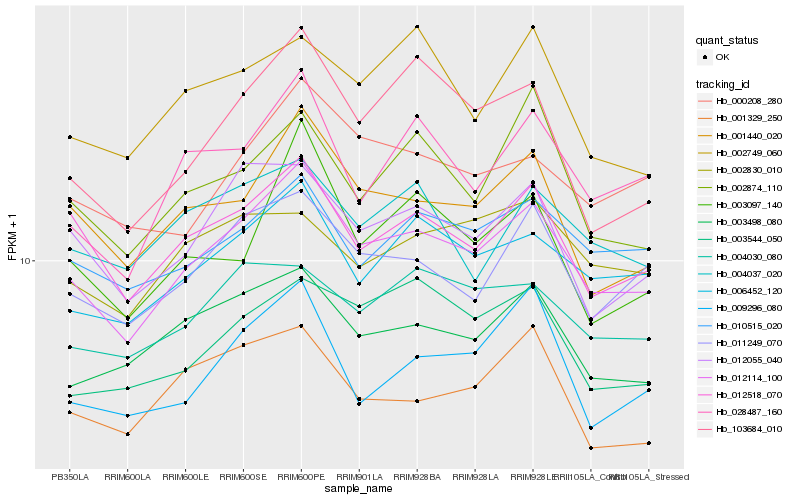
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_012114_100 |
0.0 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase At5g41260 [Jatropha curcas] |
| 2 |
Hb_012518_070 |
0.0617707314 |
- |
- |
PREDICTED: ubiquitin carboxyl-terminal hydrolase 9 isoform X1 [Jatropha curcas] |
| 3 |
Hb_006452_120 |
0.0624114824 |
- |
- |
PREDICTED: protein FAM188A [Jatropha curcas] |
| 4 |
Hb_001440_020 |
0.063073553 |
- |
- |
exocyst componenet sec8, putative [Ricinus communis] |
| 5 |
Hb_011249_070 |
0.0641390414 |
- |
- |
hypothetical protein POPTR_0018s12610g [Populus trichocarpa] |
| 6 |
Hb_012055_040 |
0.0686144935 |
- |
- |
PREDICTED: U-box domain-containing protein 4 [Jatropha curcas] |
| 7 |
Hb_003498_080 |
0.0686501872 |
- |
- |
PREDICTED: decapping 5-like protein isoform X2 [Jatropha curcas] |
| 8 |
Hb_010515_020 |
0.0715568167 |
- |
- |
PREDICTED: protein transport protein SEC23-like [Jatropha curcas] |
| 9 |
Hb_028487_160 |
0.0727849833 |
- |
- |
clathrin assembly protein, putative [Ricinus communis] |
| 10 |
Hb_000208_280 |
0.0742095418 |
transcription factor |
TF Family: NAC |
PREDICTED: NAC domain-containing protein 78 [Jatropha curcas] |
| 11 |
Hb_103684_010 |
0.0777327701 |
- |
- |
PREDICTED: cullin-1 isoform X3 [Nicotiana tomentosiformis] |
| 12 |
Hb_004030_080 |
0.0779987701 |
- |
- |
hypothetical protein JCGZ_25110 [Jatropha curcas] |
| 13 |
Hb_002874_110 |
0.0788845776 |
- |
- |
PREDICTED: uncharacterized protein LOC105637451 [Jatropha curcas] |
| 14 |
Hb_003097_140 |
0.0790448625 |
- |
- |
PREDICTED: glycosyltransferase-like KOBITO 1 [Jatropha curcas] |
| 15 |
Hb_001329_250 |
0.079626587 |
transcription factor |
TF Family: C2H2 |
PREDICTED: uncharacterized protein LOC105642692 isoform X1 [Jatropha curcas] |
| 16 |
Hb_004037_020 |
0.0800616785 |
- |
- |
PREDICTED: uncharacterized protein LOC105632366 isoform X1 [Jatropha curcas] |
| 17 |
Hb_003544_050 |
0.0811844501 |
- |
- |
PREDICTED: uncharacterized protein LOC105644300 isoform X1 [Jatropha curcas] |
| 18 |
Hb_009296_080 |
0.0816848371 |
- |
- |
PREDICTED: probable apyrase 7 isoform X1 [Jatropha curcas] |
| 19 |
Hb_002749_060 |
0.0825806735 |
- |
- |
PREDICTED: protein transport protein Sec61 subunit alpha-like [Jatropha curcas] |
| 20 |
Hb_002830_010 |
0.083473046 |
- |
- |
PREDICTED: TBCC domain-containing protein 1 [Jatropha curcas] |