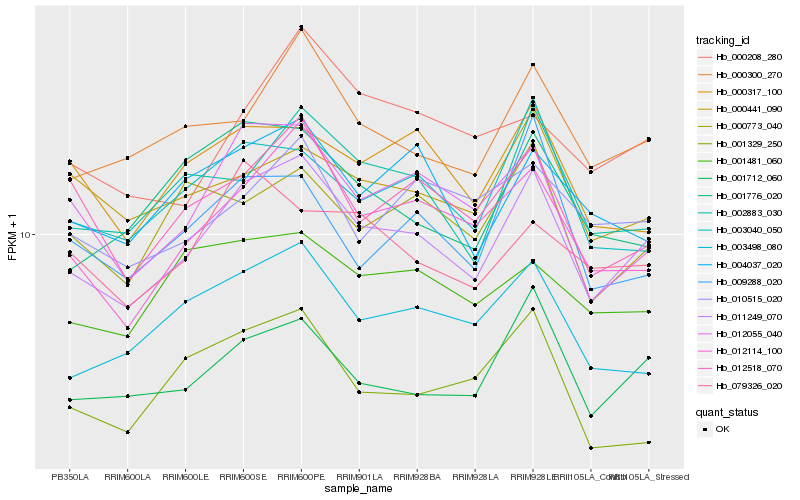
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_011249_070 |
0.0 |
- |
- |
hypothetical protein POPTR_0018s12610g [Populus trichocarpa] |
| 2 |
Hb_001712_060 |
0.0632075706 |
- |
- |
PREDICTED: proline-rich receptor-like protein kinase PERK9 [Jatropha curcas] |
| 3 |
Hb_012114_100 |
0.0641390414 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase At5g41260 [Jatropha curcas] |
| 4 |
Hb_000300_270 |
0.0673858544 |
- |
- |
cationic amino acid transporter, putative [Ricinus communis] |
| 5 |
Hb_012055_040 |
0.0680627472 |
- |
- |
PREDICTED: U-box domain-containing protein 4 [Jatropha curcas] |
| 6 |
Hb_002883_030 |
0.0708784267 |
- |
- |
PREDICTED: uncharacterized protein LOC105638222 [Jatropha curcas] |
| 7 |
Hb_003498_080 |
0.0719990355 |
- |
- |
PREDICTED: decapping 5-like protein isoform X2 [Jatropha curcas] |
| 8 |
Hb_003040_050 |
0.0735503129 |
- |
- |
PREDICTED: uncharacterized protein LOC105638219 [Jatropha curcas] |
| 9 |
Hb_012518_070 |
0.07433532 |
- |
- |
PREDICTED: ubiquitin carboxyl-terminal hydrolase 9 isoform X1 [Jatropha curcas] |
| 10 |
Hb_004037_020 |
0.0781834193 |
- |
- |
PREDICTED: uncharacterized protein LOC105632366 isoform X1 [Jatropha curcas] |
| 11 |
Hb_000441_090 |
0.0811552769 |
- |
- |
PREDICTED: serine/threonine-protein phosphatase 6 regulatory subunit 2 [Jatropha curcas] |
| 12 |
Hb_000773_040 |
0.0812339905 |
- |
- |
PREDICTED: uncharacterized protein LOC105641863 isoform X2 [Jatropha curcas] |
| 13 |
Hb_001481_060 |
0.0813974795 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase HOS1 isoform X1 [Jatropha curcas] |
| 14 |
Hb_000317_100 |
0.0814590442 |
- |
- |
PREDICTED: uncharacterized protein LOC105646995 [Jatropha curcas] |
| 15 |
Hb_001776_020 |
0.0822561011 |
- |
- |
Protein kinase capable of phosphorylating tyrosine family protein [Populus trichocarpa] |
| 16 |
Hb_009288_020 |
0.083478309 |
- |
- |
PREDICTED: protein FRIGIDA [Jatropha curcas] |
| 17 |
Hb_001329_250 |
0.084018961 |
transcription factor |
TF Family: C2H2 |
PREDICTED: uncharacterized protein LOC105642692 isoform X1 [Jatropha curcas] |
| 18 |
Hb_000208_280 |
0.0848373204 |
transcription factor |
TF Family: NAC |
PREDICTED: NAC domain-containing protein 78 [Jatropha curcas] |
| 19 |
Hb_010515_020 |
0.0851566788 |
- |
- |
PREDICTED: protein transport protein SEC23-like [Jatropha curcas] |
| 20 |
Hb_079326_020 |
0.0852615593 |
- |
- |
conserved hypothetical protein [Ricinus communis] |