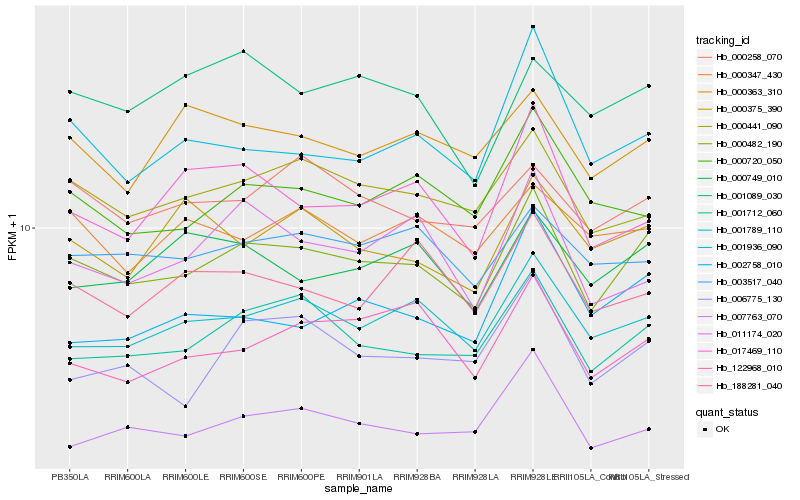
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000482_190 |
0.0 |
- |
- |
PREDICTED: serine/threonine-protein kinase EDR1 [Jatropha curcas] |
| 2 |
Hb_001712_060 |
0.0639703507 |
- |
- |
PREDICTED: proline-rich receptor-like protein kinase PERK9 [Jatropha curcas] |
| 3 |
Hb_001789_110 |
0.0681515888 |
- |
- |
ubiquitin-protein ligase, putative [Ricinus communis] |
| 4 |
Hb_000441_090 |
0.0716041363 |
- |
- |
PREDICTED: serine/threonine-protein phosphatase 6 regulatory subunit 2 [Jatropha curcas] |
| 5 |
Hb_003517_040 |
0.0809143286 |
- |
- |
PREDICTED: putative DUF21 domain-containing protein At3g13070, chloroplastic [Jatropha curcas] |
| 6 |
Hb_001936_090 |
0.082767277 |
- |
- |
PREDICTED: BOI-related E3 ubiquitin-protein ligase 1-like isoform X1 [Jatropha curcas] |
| 7 |
Hb_000258_070 |
0.0829395667 |
- |
- |
PREDICTED: uncharacterized protein LOC105649104 [Jatropha curcas] |
| 8 |
Hb_000749_010 |
0.0834935045 |
- |
- |
PREDICTED: non-canonical poly(A) RNA polymerase PAPD5 isoform X1 [Jatropha curcas] |
| 9 |
Hb_000720_050 |
0.0839782868 |
- |
- |
PREDICTED: uncharacterized protein LOC105645857 isoform X1 [Jatropha curcas] |
| 10 |
Hb_007763_070 |
0.0846249058 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g71460, chloroplastic [Jatropha curcas] |
| 11 |
Hb_122968_010 |
0.0856506123 |
- |
- |
hypothetical protein JCGZ_23576 [Jatropha curcas] |
| 12 |
Hb_000363_310 |
0.0858314688 |
- |
- |
DNA topoisomerase type I, putative [Ricinus communis] |
| 13 |
Hb_006775_130 |
0.086966501 |
- |
- |
PREDICTED: bidirectional sugar transporter SWEET2a-like [Jatropha curcas] |
| 14 |
Hb_001089_030 |
0.0872321948 |
- |
- |
PREDICTED: uncharacterized protein LOC105638026 [Jatropha curcas] |
| 15 |
Hb_188281_040 |
0.0888220004 |
- |
- |
PREDICTED: transcription initiation factor TFIID subunit 12 [Jatropha curcas] |
| 16 |
Hb_017469_110 |
0.088868112 |
- |
- |
PREDICTED: uncharacterized protein LOC105641795 [Jatropha curcas] |
| 17 |
Hb_000347_430 |
0.0890828703 |
- |
- |
PREDICTED: mitochondrial ubiquitin ligase activator of nfkb 1-A [Jatropha curcas] |
| 18 |
Hb_000375_390 |
0.0890906341 |
- |
- |
poly-A binding protein, putative [Ricinus communis] |
| 19 |
Hb_011174_020 |
0.0908506339 |
- |
- |
PREDICTED: uncharacterized protein LOC105632610 [Jatropha curcas] |
| 20 |
Hb_002758_010 |
0.091016464 |
- |
- |
PREDICTED: ribonuclease II, chloroplastic/mitochondrial [Populus euphratica] |