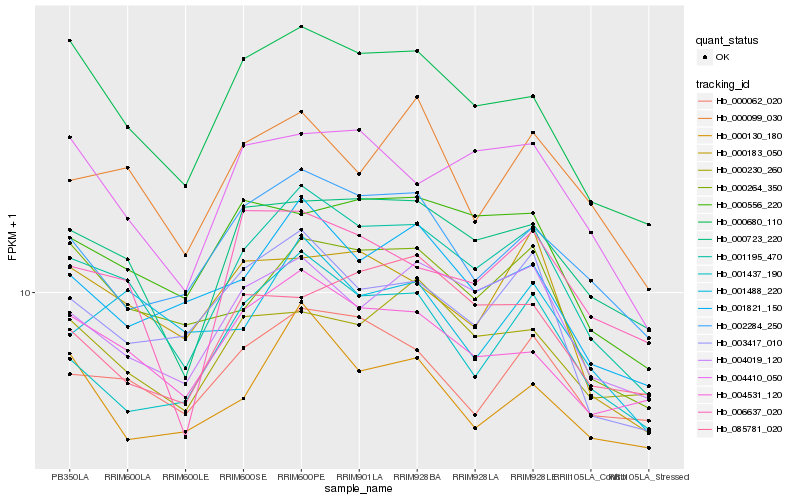
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001195_470 |
0.0 |
- |
- |
PREDICTED: uncharacterized membrane protein At1g75140 [Jatropha curcas] |
| 2 |
Hb_000723_220 |
0.0705426529 |
- |
- |
PREDICTED: uncharacterized protein LOC105642234 [Jatropha curcas] |
| 3 |
Hb_004019_120 |
0.0736567134 |
- |
- |
PREDICTED: conserved oligomeric Golgi complex subunit 1 [Jatropha curcas] |
| 4 |
Hb_000680_110 |
0.0757848215 |
- |
- |
Endosomal P24A protein precursor, putative [Ricinus communis] |
| 5 |
Hb_003417_010 |
0.0827924591 |
- |
- |
PREDICTED: protein TPLATE [Jatropha curcas] |
| 6 |
Hb_001821_150 |
0.085101388 |
- |
- |
PREDICTED: potassium transporter 7-like [Jatropha curcas] |
| 7 |
Hb_004531_120 |
0.085201635 |
- |
- |
PREDICTED: uncharacterized protein LOC105635025 [Jatropha curcas] |
| 8 |
Hb_001437_190 |
0.0856956868 |
- |
- |
ABC transporter family protein [Hevea brasiliensis] |
| 9 |
Hb_000556_220 |
0.0857417582 |
- |
- |
PREDICTED: brefeldin A-inhibited guanine nucleotide-exchange protein 2-like [Jatropha curcas] |
| 10 |
Hb_000264_350 |
0.086313693 |
- |
- |
PREDICTED: uncharacterized protein LOC105649541 [Jatropha curcas] |
| 11 |
Hb_002284_250 |
0.0866522924 |
- |
- |
PREDICTED: epidermal growth factor receptor substrate 15-like 1 [Jatropha curcas] |
| 12 |
Hb_004410_050 |
0.0884005165 |
- |
- |
PREDICTED: uncharacterized TPR repeat-containing protein At1g05150-like [Jatropha curcas] |
| 13 |
Hb_000062_020 |
0.0897224159 |
- |
- |
ubiquitin-protein ligase, putative [Ricinus communis] |
| 14 |
Hb_000099_030 |
0.090470134 |
- |
- |
6-phosphogluconate dehydrogenase, putative [Ricinus communis] |
| 15 |
Hb_006637_020 |
0.0948856934 |
- |
- |
PREDICTED: uncharacterized protein LOC105642234 [Jatropha curcas] |
| 16 |
Hb_000183_050 |
0.0958066148 |
- |
- |
PREDICTED: alpha-mannosidase 2 [Jatropha curcas] |
| 17 |
Hb_000230_260 |
0.0992497622 |
- |
- |
PREDICTED: putative E3 ubiquitin-protein ligase RF298 [Jatropha curcas] |
| 18 |
Hb_001488_220 |
0.1021591713 |
- |
- |
PREDICTED: exocyst complex component EXO70B1 [Jatropha curcas] |
| 19 |
Hb_085781_020 |
0.1023129035 |
- |
- |
PREDICTED: extra-large guanine nucleotide-binding protein 1 [Jatropha curcas] |
| 20 |
Hb_000130_180 |
0.1047021083 |
- |
- |
PREDICTED: GPI inositol-deacylase-like isoform X2 [Jatropha curcas] |