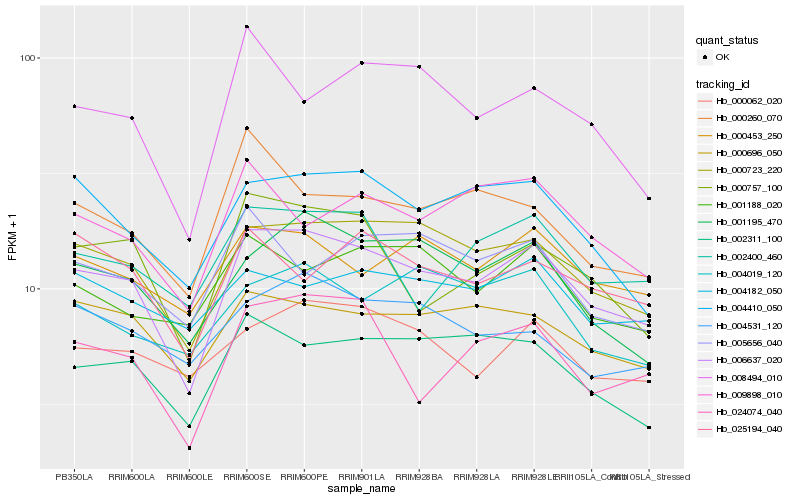
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_006637_020 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105642234 [Jatropha curcas] |
| 2 |
Hb_000723_220 |
0.0680412754 |
- |
- |
PREDICTED: uncharacterized protein LOC105642234 [Jatropha curcas] |
| 3 |
Hb_002311_100 |
0.0923179416 |
transcription factor |
TF Family: FAR1 |
PREDICTED: protein FAR1-RELATED SEQUENCE 11-like [Jatropha curcas] |
| 4 |
Hb_000696_050 |
0.0942143419 |
- |
- |
PREDICTED: CSC1-like protein At4g35870 [Jatropha curcas] |
| 5 |
Hb_001195_470 |
0.0948856934 |
- |
- |
PREDICTED: uncharacterized membrane protein At1g75140 [Jatropha curcas] |
| 6 |
Hb_004410_050 |
0.0977444491 |
- |
- |
PREDICTED: uncharacterized TPR repeat-containing protein At1g05150-like [Jatropha curcas] |
| 7 |
Hb_000757_100 |
0.0980923799 |
- |
- |
SPla/RYanodine receptor domain-containing protein isoform 2 [Theobroma cacao] |
| 8 |
Hb_000453_250 |
0.1011571874 |
- |
- |
PREDICTED: uncharacterized protein LOC105637185 [Jatropha curcas] |
| 9 |
Hb_024074_040 |
0.1018975696 |
- |
- |
PREDICTED: leucine--tRNA ligase, cytoplasmic [Jatropha curcas] |
| 10 |
Hb_000062_020 |
0.1046111173 |
- |
- |
ubiquitin-protein ligase, putative [Ricinus communis] |
| 11 |
Hb_002400_460 |
0.1063658395 |
- |
- |
PREDICTED: uncharacterized protein LOC105637599 isoform X2 [Jatropha curcas] |
| 12 |
Hb_009898_010 |
0.1073947135 |
transcription factor |
TF Family: MYB |
PREDICTED: uncharacterized protein LOC105645502 [Jatropha curcas] |
| 13 |
Hb_008494_010 |
0.1074769381 |
- |
- |
PREDICTED: uncharacterized protein LOC105636015 [Jatropha curcas] |
| 14 |
Hb_004531_120 |
0.109123602 |
- |
- |
PREDICTED: uncharacterized protein LOC105635025 [Jatropha curcas] |
| 15 |
Hb_025194_040 |
0.1093324168 |
- |
- |
PREDICTED: uncharacterized protein LOC105644854 isoform X2 [Jatropha curcas] |
| 16 |
Hb_001188_020 |
0.109682258 |
- |
- |
PREDICTED: E3 SUMO-protein ligase SIZ1 isoform X1 [Jatropha curcas] |
| 17 |
Hb_000260_070 |
0.1104787495 |
- |
- |
kinase, putative [Ricinus communis] |
| 18 |
Hb_004019_120 |
0.1120627442 |
- |
- |
PREDICTED: conserved oligomeric Golgi complex subunit 1 [Jatropha curcas] |
| 19 |
Hb_004182_050 |
0.1126203756 |
- |
- |
PREDICTED: serine/threonine-protein phosphatase BSL3 isoform X2 [Jatropha curcas] |
| 20 |
Hb_005656_040 |
0.1132045071 |
desease resistance |
Gene Name: T2SE |
ATP-dependent RNA helicase, putative [Ricinus communis] |