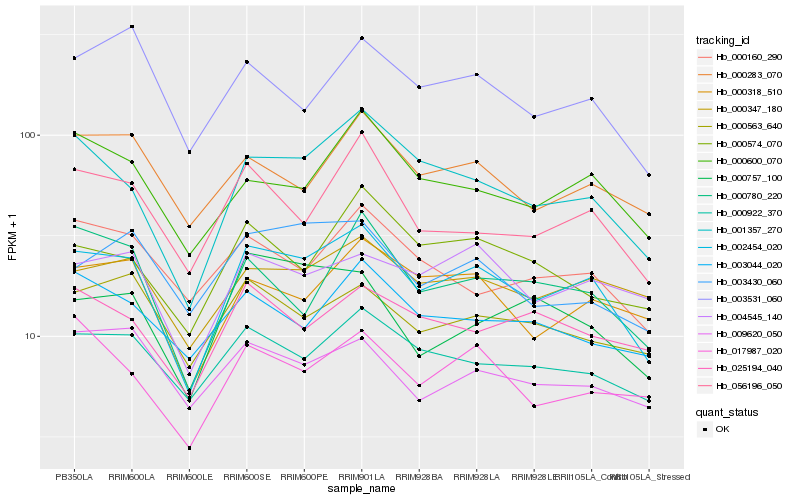
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002454_020 |
0.0 |
transcription factor |
TF Family: HB |
PREDICTED: BEL1-like homeodomain protein 9 [Vitis vinifera] |
| 2 |
Hb_001357_270 |
0.0874965835 |
- |
- |
PREDICTED: aspartic proteinase A1-like [Jatropha curcas] |
| 3 |
Hb_000922_370 |
0.0986567238 |
- |
- |
PREDICTED: uncharacterized protein LOC105640366 [Jatropha curcas] |
| 4 |
Hb_000318_510 |
0.1022502634 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At2g15980 [Jatropha curcas] |
| 5 |
Hb_000347_180 |
0.1046841091 |
- |
- |
PREDICTED: exocyst complex component EXO70A1-like [Jatropha curcas] |
| 6 |
Hb_056196_050 |
0.1051043643 |
- |
- |
PREDICTED: nuclear poly(A) polymerase 4-like [Jatropha curcas] |
| 7 |
Hb_000600_070 |
0.1053742544 |
- |
- |
PREDICTED: leucine aminopeptidase 1-like [Jatropha curcas] |
| 8 |
Hb_004545_140 |
0.1087860524 |
- |
- |
hypothetical protein JCGZ_23405 [Jatropha curcas] |
| 9 |
Hb_000780_220 |
0.1094722932 |
transcription factor |
TF Family: GRAS |
PREDICTED: scarecrow-like transcription factor PAT1 [Jatropha curcas] |
| 10 |
Hb_000283_070 |
0.109914315 |
- |
- |
PREDICTED: T-complex protein 1 subunit delta [Jatropha curcas] |
| 11 |
Hb_003531_060 |
0.1125060842 |
- |
- |
unnamed protein product [Coffea canephora] |
| 12 |
Hb_000563_640 |
0.1134084901 |
- |
- |
PREDICTED: chaperone protein dnaJ 49 [Jatropha curcas] |
| 13 |
Hb_003430_060 |
0.1136529165 |
- |
- |
PREDICTED: potassium transporter 23 isoform X2 [Elaeis guineensis] |
| 14 |
Hb_017987_020 |
0.1142466024 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_000574_070 |
0.1154905061 |
- |
- |
PREDICTED: DEAD-box ATP-dependent RNA helicase 35 [Jatropha curcas] |
| 16 |
Hb_009620_050 |
0.1158423422 |
- |
- |
PREDICTED: protein unc-45 homolog A [Jatropha curcas] |
| 17 |
Hb_025194_040 |
0.1160617953 |
- |
- |
PREDICTED: uncharacterized protein LOC105644854 isoform X2 [Jatropha curcas] |
| 18 |
Hb_000757_100 |
0.1179087419 |
- |
- |
SPla/RYanodine receptor domain-containing protein isoform 2 [Theobroma cacao] |
| 19 |
Hb_000160_290 |
0.1180699363 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 20 |
Hb_003044_020 |
0.1183674162 |
- |
- |
PREDICTED: uncharacterized protein LOC105641479 [Jatropha curcas] |