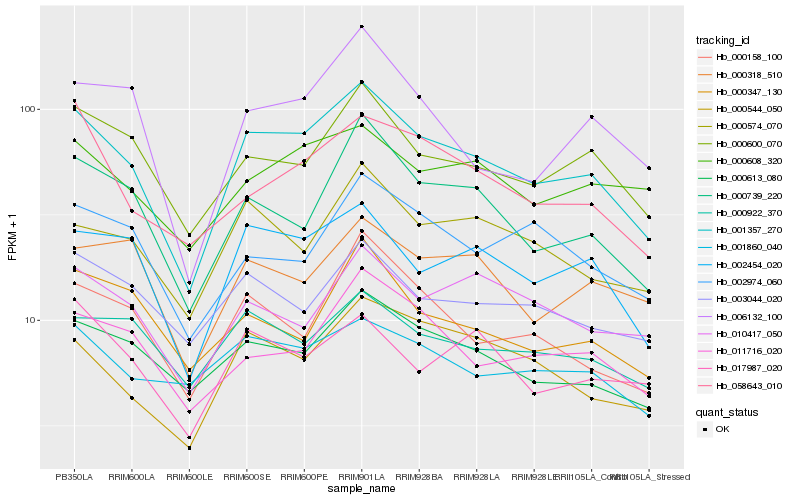
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001357_270 |
0.0 |
- |
- |
PREDICTED: aspartic proteinase A1-like [Jatropha curcas] |
| 2 |
Hb_002454_020 |
0.0874965835 |
transcription factor |
TF Family: HB |
PREDICTED: BEL1-like homeodomain protein 9 [Vitis vinifera] |
| 3 |
Hb_000739_220 |
0.0957008708 |
- |
- |
PREDICTED: brefeldin A-inhibited guanine nucleotide-exchange protein 2-like [Jatropha curcas] |
| 4 |
Hb_000600_070 |
0.0962587188 |
- |
- |
PREDICTED: leucine aminopeptidase 1-like [Jatropha curcas] |
| 5 |
Hb_000613_080 |
0.0970527493 |
- |
- |
PREDICTED: mucin-5AC-like [Nicotiana sylvestris] |
| 6 |
Hb_000544_050 |
0.1027888852 |
- |
- |
PREDICTED: serine/threonine-protein kinase EDR1 [Jatropha curcas] |
| 7 |
Hb_000158_100 |
0.1047634696 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_011716_020 |
0.1059579145 |
- |
- |
PREDICTED: uncharacterized protein LOC105637783 [Jatropha curcas] |
| 9 |
Hb_000574_070 |
0.1067379002 |
- |
- |
PREDICTED: DEAD-box ATP-dependent RNA helicase 35 [Jatropha curcas] |
| 10 |
Hb_006132_100 |
0.1117188533 |
- |
- |
PREDICTED: uncharacterized protein LOC105642968 [Jatropha curcas] |
| 11 |
Hb_058643_010 |
0.1120976971 |
- |
- |
hydrolase, putative [Ricinus communis] |
| 12 |
Hb_003044_020 |
0.1126403648 |
- |
- |
PREDICTED: uncharacterized protein LOC105641479 [Jatropha curcas] |
| 13 |
Hb_000922_370 |
0.1132155159 |
- |
- |
PREDICTED: uncharacterized protein LOC105640366 [Jatropha curcas] |
| 14 |
Hb_001860_040 |
0.1152593805 |
- |
- |
PREDICTED: cleavage stimulation factor subunit 77 isoform X4 [Jatropha curcas] |
| 15 |
Hb_000608_320 |
0.1158532192 |
- |
- |
PREDICTED: uncharacterized protein LOC105644891 [Jatropha curcas] |
| 16 |
Hb_017987_020 |
0.1160702123 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_002974_060 |
0.1172637517 |
- |
- |
PREDICTED: transcription initiation factor TFIID subunit 15b isoform X2 [Jatropha curcas] |
| 18 |
Hb_000318_510 |
0.1173684388 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At2g15980 [Jatropha curcas] |
| 19 |
Hb_010417_050 |
0.1178495929 |
- |
- |
PREDICTED: uncharacterized protein LOC105645746 [Jatropha curcas] |
| 20 |
Hb_000347_130 |
0.1182794132 |
- |
- |
PREDICTED: nuclear pore complex protein NUP107 [Jatropha curcas] |