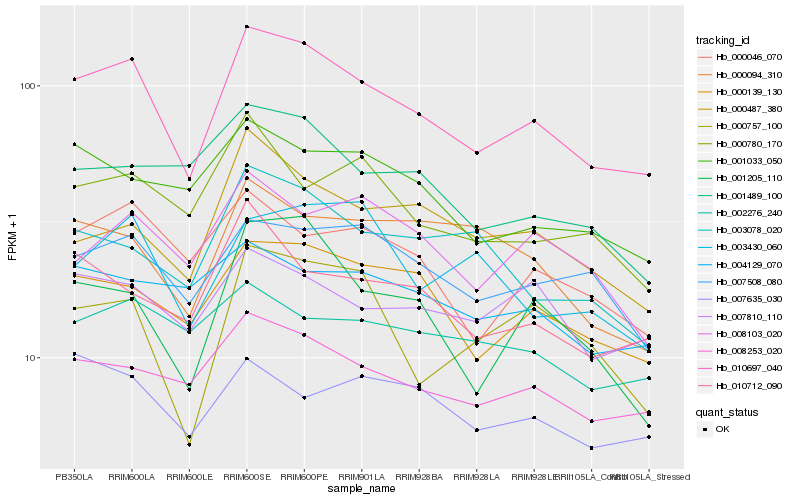
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_010697_040 |
0.0 |
- |
- |
PREDICTED: protein GPR107 isoform X1 [Jatropha curcas] |
| 2 |
Hb_000139_130 |
0.0787447001 |
- |
- |
PREDICTED: chloride channel protein CLC-d isoform X1 [Jatropha curcas] |
| 3 |
Hb_008253_020 |
0.0837606908 |
- |
- |
PREDICTED: serine/threonine protein phosphatase 2A 57 kDa regulatory subunit B' theta isoform-like [Jatropha curcas] |
| 4 |
Hb_007508_080 |
0.0893517937 |
- |
- |
PREDICTED: presenilin-like protein At2g29900 [Jatropha curcas] |
| 5 |
Hb_001205_110 |
0.0911727049 |
transcription factor |
TF Family: GRAS |
PREDICTED: scarecrow-like protein 4 [Jatropha curcas] |
| 6 |
Hb_000046_070 |
0.0929187268 |
transcription factor |
TF Family: SBP |
Squamosa promoter-binding protein, putative [Ricinus communis] |
| 7 |
Hb_010712_090 |
0.0945233364 |
- |
- |
PREDICTED: uncharacterized protein LOC105631276 [Jatropha curcas] |
| 8 |
Hb_000487_380 |
0.0957860551 |
- |
- |
PREDICTED: protein PAT1 homolog 1 isoform X1 [Jatropha curcas] |
| 9 |
Hb_001033_050 |
0.096452584 |
- |
- |
PREDICTED: acetylornithine deacetylase [Jatropha curcas] |
| 10 |
Hb_007810_110 |
0.0973187331 |
- |
- |
PREDICTED: DNA/RNA-binding protein KIN17 [Jatropha curcas] |
| 11 |
Hb_000780_170 |
0.1000316487 |
- |
- |
PREDICTED: OBERON-like protein [Jatropha curcas] |
| 12 |
Hb_003078_020 |
0.1015995715 |
- |
- |
PREDICTED: U-box domain-containing protein 9 [Jatropha curcas] |
| 13 |
Hb_008103_020 |
0.1016852566 |
- |
- |
hypothetical protein POPTR_0003s20310g [Populus trichocarpa] |
| 14 |
Hb_001489_100 |
0.1031100169 |
- |
- |
PREDICTED: hypersensitive-induced response protein 2 [Jatropha curcas] |
| 15 |
Hb_007635_030 |
0.1031834942 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g77360, mitochondrial-like [Jatropha curcas] |
| 16 |
Hb_000757_100 |
0.1038352347 |
- |
- |
SPla/RYanodine receptor domain-containing protein isoform 2 [Theobroma cacao] |
| 17 |
Hb_000094_310 |
0.105045602 |
- |
- |
PREDICTED: uncharacterized membrane protein At1g75140 [Jatropha curcas] |
| 18 |
Hb_004129_070 |
0.1052192589 |
- |
- |
PREDICTED: conserved oligomeric Golgi complex subunit 5 [Jatropha curcas] |
| 19 |
Hb_003430_060 |
0.1064148618 |
- |
- |
PREDICTED: potassium transporter 23 isoform X2 [Elaeis guineensis] |
| 20 |
Hb_002276_240 |
0.1067002391 |
- |
- |
PREDICTED: uncharacterized protein LOC105640682 [Jatropha curcas] |