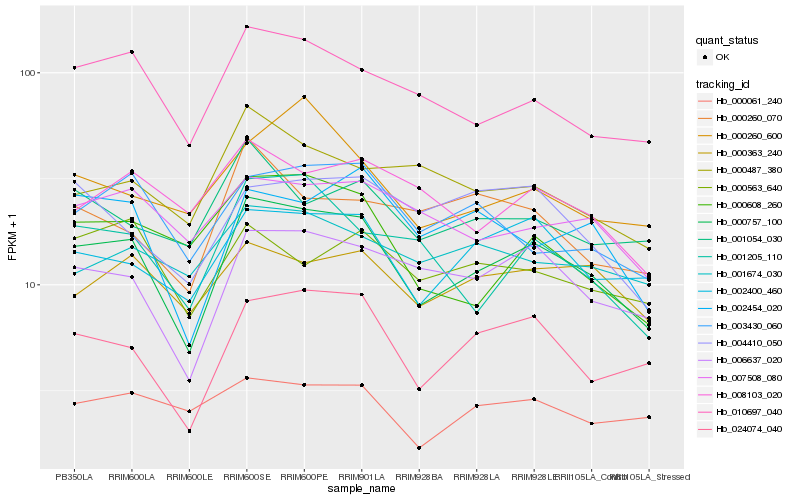
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000757_100 |
0.0 |
- |
- |
SPla/RYanodine receptor domain-containing protein isoform 2 [Theobroma cacao] |
| 2 |
Hb_024074_040 |
0.0949986128 |
- |
- |
PREDICTED: leucine--tRNA ligase, cytoplasmic [Jatropha curcas] |
| 3 |
Hb_006637_020 |
0.0980923799 |
- |
- |
PREDICTED: uncharacterized protein LOC105642234 [Jatropha curcas] |
| 4 |
Hb_010697_040 |
0.1038352347 |
- |
- |
PREDICTED: protein GPR107 isoform X1 [Jatropha curcas] |
| 5 |
Hb_002400_460 |
0.1067714366 |
- |
- |
PREDICTED: uncharacterized protein LOC105637599 isoform X2 [Jatropha curcas] |
| 6 |
Hb_002454_020 |
0.1179087419 |
transcription factor |
TF Family: HB |
PREDICTED: BEL1-like homeodomain protein 9 [Vitis vinifera] |
| 7 |
Hb_003430_060 |
0.1213512028 |
- |
- |
PREDICTED: potassium transporter 23 isoform X2 [Elaeis guineensis] |
| 8 |
Hb_001205_110 |
0.1221017143 |
transcription factor |
TF Family: GRAS |
PREDICTED: scarecrow-like protein 4 [Jatropha curcas] |
| 9 |
Hb_000363_240 |
0.1230068335 |
- |
- |
PREDICTED: beta-(1,2)-xylosyltransferase [Jatropha curcas] |
| 10 |
Hb_000608_260 |
0.1245428557 |
- |
- |
PREDICTED: probable protein phosphatase 2C 33 [Jatropha curcas] |
| 11 |
Hb_007508_080 |
0.1247662297 |
- |
- |
PREDICTED: presenilin-like protein At2g29900 [Jatropha curcas] |
| 12 |
Hb_001054_030 |
0.1298509344 |
transcription factor |
TF Family: NF-X1 |
PREDICTED: NF-X1-type zinc finger protein NFXL1 [Jatropha curcas] |
| 13 |
Hb_000563_640 |
0.1305035195 |
- |
- |
PREDICTED: chaperone protein dnaJ 49 [Jatropha curcas] |
| 14 |
Hb_000487_380 |
0.1310270147 |
- |
- |
PREDICTED: protein PAT1 homolog 1 isoform X1 [Jatropha curcas] |
| 15 |
Hb_008103_020 |
0.1314825555 |
- |
- |
hypothetical protein POPTR_0003s20310g [Populus trichocarpa] |
| 16 |
Hb_001674_030 |
0.1328617043 |
- |
- |
PREDICTED: BTB/POZ domain-containing protein At2g24240 [Jatropha curcas] |
| 17 |
Hb_000260_600 |
0.1331469057 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_004410_050 |
0.1331929459 |
- |
- |
PREDICTED: uncharacterized TPR repeat-containing protein At1g05150-like [Jatropha curcas] |
| 19 |
Hb_000260_070 |
0.1343650723 |
- |
- |
kinase, putative [Ricinus communis] |
| 20 |
Hb_000061_240 |
0.1347824007 |
- |
- |
kinase, putative [Ricinus communis] |