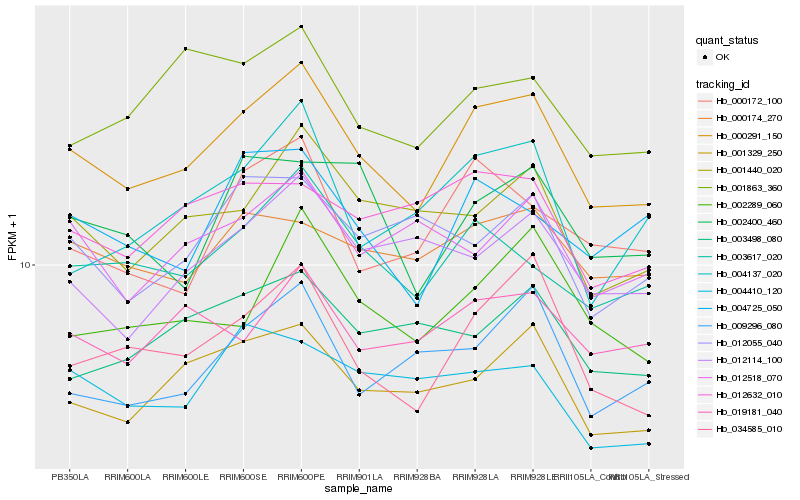
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000291_150 |
0.0 |
- |
- |
PREDICTED: serine hydroxymethyltransferase 7 [Jatropha curcas] |
| 2 |
Hb_034585_010 |
0.0805819562 |
- |
- |
PREDICTED: uncharacterized protein LOC105639637 [Jatropha curcas] |
| 3 |
Hb_003617_020 |
0.0829896405 |
- |
- |
PREDICTED: uncharacterized protein LOC105638179 [Jatropha curcas] |
| 4 |
Hb_001329_250 |
0.0842861148 |
transcription factor |
TF Family: C2H2 |
PREDICTED: uncharacterized protein LOC105642692 isoform X1 [Jatropha curcas] |
| 5 |
Hb_001440_020 |
0.0902830893 |
- |
- |
exocyst componenet sec8, putative [Ricinus communis] |
| 6 |
Hb_004725_050 |
0.0947385289 |
- |
- |
Rhomboid protein, putative [Ricinus communis] |
| 7 |
Hb_002289_060 |
0.0951203699 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_000174_270 |
0.0956662191 |
- |
- |
PREDICTED: type II inositol 1,4,5-trisphosphate 5-phosphatase FRA3 isoform X1 [Jatropha curcas] |
| 9 |
Hb_000172_100 |
0.0963662193 |
- |
- |
PREDICTED: acyl-coenzyme A oxidase 2, peroxisomal [Jatropha curcas] |
| 10 |
Hb_019181_040 |
0.0964909966 |
- |
- |
component of oligomeric golgi complex, putative [Ricinus communis] |
| 11 |
Hb_009296_080 |
0.1007548452 |
- |
- |
PREDICTED: probable apyrase 7 isoform X1 [Jatropha curcas] |
| 12 |
Hb_002400_460 |
0.1017410415 |
- |
- |
PREDICTED: uncharacterized protein LOC105637599 isoform X2 [Jatropha curcas] |
| 13 |
Hb_012114_100 |
0.1025322277 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase At5g41260 [Jatropha curcas] |
| 14 |
Hb_001863_360 |
0.1027775115 |
- |
- |
PREDICTED: 5'-adenylylsulfate reductase-like 4 [Jatropha curcas] |
| 15 |
Hb_004137_020 |
0.1028820515 |
- |
- |
PREDICTED: putative UDP-glucuronate:xylan alpha-glucuronosyltransferase 3 isoform X2 [Jatropha curcas] |
| 16 |
Hb_012632_010 |
0.1029542349 |
- |
- |
PREDICTED: uncharacterized protein LOC105640019 [Jatropha curcas] |
| 17 |
Hb_003498_080 |
0.1049777567 |
- |
- |
PREDICTED: decapping 5-like protein isoform X2 [Jatropha curcas] |
| 18 |
Hb_012518_070 |
0.1058759129 |
- |
- |
PREDICTED: ubiquitin carboxyl-terminal hydrolase 9 isoform X1 [Jatropha curcas] |
| 19 |
Hb_004410_120 |
0.106976504 |
- |
- |
PREDICTED: uncharacterized protein LOC105633877 isoform X1 [Jatropha curcas] |
| 20 |
Hb_012055_040 |
0.1094343303 |
- |
- |
PREDICTED: U-box domain-containing protein 4 [Jatropha curcas] |