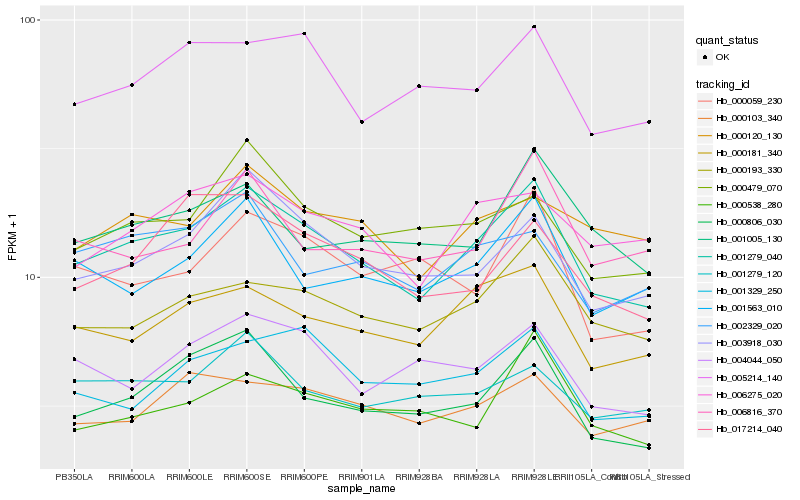
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001279_040 |
0.0 |
- |
- |
PREDICTED: probable carboxylesterase 11 [Jatropha curcas] |
| 2 |
Hb_000806_030 |
0.0747504686 |
transcription factor |
TF Family: C2H2 |
PREDICTED: probable lysine-specific demethylase ELF6 [Jatropha curcas] |
| 3 |
Hb_000181_340 |
0.0777902244 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase At1g54610 [Jatropha curcas] |
| 4 |
Hb_000193_330 |
0.0792452348 |
- |
- |
PREDICTED: formin-like protein 20 [Jatropha curcas] |
| 5 |
Hb_003918_030 |
0.082080926 |
- |
- |
PREDICTED: oxysterol-binding protein-related protein 1C-like isoform X1 [Jatropha curcas] |
| 6 |
Hb_005214_140 |
0.0872911522 |
- |
- |
PREDICTED: transcription factor GTE2-like [Jatropha curcas] |
| 7 |
Hb_001563_010 |
0.0890921143 |
transcription factor |
TF Family: SET |
PREDICTED: probable histone-lysine N-methyltransferase ATXR3 [Jatropha curcas] |
| 8 |
Hb_006275_020 |
0.0896798212 |
- |
- |
PREDICTED: VIN3-like protein 2 [Jatropha curcas] |
| 9 |
Hb_000479_070 |
0.0907401045 |
- |
- |
PREDICTED: uncharacterized protein LOC105644141 [Jatropha curcas] |
| 10 |
Hb_000103_340 |
0.0926816308 |
- |
- |
PREDICTED: uncharacterized protein LOC105632051 isoform X4 [Jatropha curcas] |
| 11 |
Hb_004044_050 |
0.0932473222 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 34-like [Jatropha curcas] |
| 12 |
Hb_001005_130 |
0.0934535134 |
- |
- |
PREDICTED: uncharacterized protein LOC105630220 isoform X4 [Jatropha curcas] |
| 13 |
Hb_000538_280 |
0.093547163 |
- |
- |
PREDICTED: uncharacterized protein LOC105642112 [Jatropha curcas] |
| 14 |
Hb_001329_250 |
0.0937974271 |
transcription factor |
TF Family: C2H2 |
PREDICTED: uncharacterized protein LOC105642692 isoform X1 [Jatropha curcas] |
| 15 |
Hb_002329_020 |
0.0948141085 |
- |
- |
PREDICTED: uncharacterized protein LOC105648015 [Jatropha curcas] |
| 16 |
Hb_017214_040 |
0.0953412417 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase COP1 isoform X1 [Jatropha curcas] |
| 17 |
Hb_006816_370 |
0.0953660102 |
transcription factor |
TF Family: PHD |
PREDICTED: uncharacterized protein LOC105647456 [Jatropha curcas] |
| 18 |
Hb_001279_120 |
0.0956275049 |
transcription factor |
TF Family: FAR1 |
PREDICTED: protein FAR1-RELATED SEQUENCE 5-like [Jatropha curcas] |
| 19 |
Hb_000120_130 |
0.0957749482 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 66 [Jatropha curcas] |
| 20 |
Hb_000059_230 |
0.098674712 |
- |
- |
PREDICTED: uncharacterized protein LOC105648049 [Jatropha curcas] |