| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
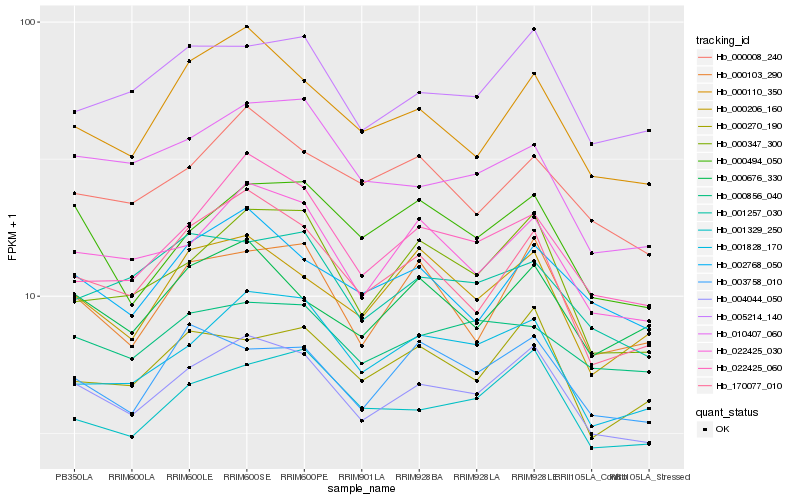
Hb_004044_050 |
0.0 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 34-like [Jatropha curcas] |
| 2 |
Hb_022425_030 |
0.0574869358 |
- |
- |
PREDICTED: casein kinase I [Jatropha curcas] |
| 3 |
Hb_005214_140 |
0.06433335 |
- |
- |
PREDICTED: transcription factor GTE2-like [Jatropha curcas] |
| 4 |
Hb_001329_250 |
0.0666060936 |
transcription factor |
TF Family: C2H2 |
PREDICTED: uncharacterized protein LOC105642692 isoform X1 [Jatropha curcas] |
| 5 |
Hb_000347_300 |
0.0684650912 |
- |
- |
PREDICTED: cation/calcium exchanger 4-like [Jatropha curcas] |
| 6 |
Hb_000110_350 |
0.0695432096 |
- |
- |
PREDICTED: ubiquitin receptor RAD23c-like isoform X2 [Gossypium raimondii] |
| 7 |
Hb_170077_010 |
0.0700372013 |
- |
- |
PREDICTED: probable ADP-ribosylation factor GTPase-activating protein AGD14 isoform X1 [Jatropha curcas] |
| 8 |
Hb_001828_170 |
0.0700874453 |
- |
- |
PREDICTED: protein TPLATE [Jatropha curcas] |
| 9 |
Hb_002768_050 |
0.0706482266 |
- |
- |
Beta-1,3-galactosyltransferase sqv-2, putative [Ricinus communis] |
| 10 |
Hb_000103_290 |
0.072427853 |
- |
- |
PREDICTED: dymeclin isoform X1 [Jatropha curcas] |
| 11 |
Hb_000008_240 |
0.0731231099 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_022425_060 |
0.0737246995 |
- |
- |
PREDICTED: protein phosphatase 2C and cyclic nucleotide-binding/kinase domain-containing protein isoform X1 [Jatropha curcas] |
| 13 |
Hb_000676_330 |
0.0737420641 |
- |
- |
protein with unknown function [Ricinus communis] |
| 14 |
Hb_000270_190 |
0.075872351 |
transcription factor |
TF Family: DDT |
tRNA ligase, putative [Ricinus communis] |
| 15 |
Hb_000856_040 |
0.0759770989 |
transcription factor |
TF Family: DDT |
PREDICTED: uncharacterized protein LOC105640470 [Jatropha curcas] |
| 16 |
Hb_000206_160 |
0.076076395 |
- |
- |
PREDICTED: TBCC domain-containing protein 1 [Jatropha curcas] |
| 17 |
Hb_000494_050 |
0.0765906666 |
- |
- |
cationic amino acid transporter, putative [Ricinus communis] |
| 18 |
Hb_003758_010 |
0.0770814547 |
transcription factor |
TF Family: VOZ |
PREDICTED: transcription factor VOZ1 isoform X1 [Jatropha curcas] |
| 19 |
Hb_001257_030 |
0.0771481014 |
- |
- |
PREDICTED: serine/threonine-protein kinase Nek5 [Jatropha curcas] |
| 20 |
Hb_010407_060 |
0.0772151731 |
- |
- |
PREDICTED: probable methyltransferase PMT3 [Jatropha curcas] |