| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
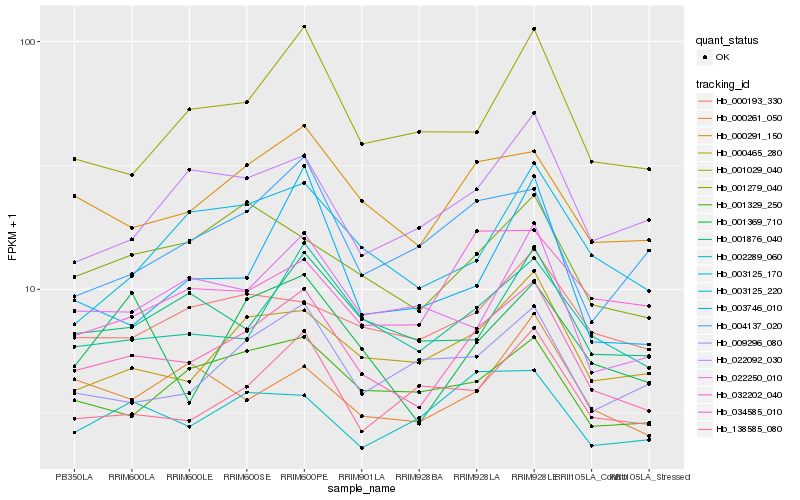
Hb_034585_010 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105639637 [Jatropha curcas] |
| 2 |
Hb_000291_150 |
0.0805819562 |
- |
- |
PREDICTED: serine hydroxymethyltransferase 7 [Jatropha curcas] |
| 3 |
Hb_002289_060 |
0.0890177981 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_000465_280 |
0.0963259966 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g47360 [Jatropha curcas] |
| 5 |
Hb_001329_250 |
0.1051528908 |
transcription factor |
TF Family: C2H2 |
PREDICTED: uncharacterized protein LOC105642692 isoform X1 [Jatropha curcas] |
| 6 |
Hb_001279_040 |
0.1054968508 |
- |
- |
PREDICTED: probable carboxylesterase 11 [Jatropha curcas] |
| 7 |
Hb_001029_040 |
0.1057126993 |
- |
- |
PREDICTED: uncharacterized protein LOC105641058 [Jatropha curcas] |
| 8 |
Hb_138585_080 |
0.1070679144 |
- |
- |
PREDICTED: uncharacterized protein LOC105648741 [Jatropha curcas] |
| 9 |
Hb_001876_040 |
0.1086794728 |
- |
- |
PREDICTED: mucin-5B [Jatropha curcas] |
| 10 |
Hb_009296_080 |
0.1125438095 |
- |
- |
PREDICTED: probable apyrase 7 isoform X1 [Jatropha curcas] |
| 11 |
Hb_022250_010 |
0.1127327606 |
- |
- |
PREDICTED: probable beta-1,4-xylosyltransferase IRX10L [Jatropha curcas] |
| 12 |
Hb_000193_330 |
0.1130059686 |
- |
- |
PREDICTED: formin-like protein 20 [Jatropha curcas] |
| 13 |
Hb_022092_030 |
0.1162334205 |
- |
- |
PREDICTED: lipoyl synthase, mitochondrial [Jatropha curcas] |
| 14 |
Hb_003125_220 |
0.1184873755 |
- |
- |
PREDICTED: CBS domain-containing protein CBSCBSPB1 [Jatropha curcas] |
| 15 |
Hb_000261_050 |
0.1195159615 |
- |
- |
PREDICTED: probable magnesium transporter NIPA2 [Populus euphratica] |
| 16 |
Hb_003125_170 |
0.1197854076 |
- |
- |
PREDICTED: uncharacterized protein LOC105648791 [Jatropha curcas] |
| 17 |
Hb_001369_710 |
0.1206754736 |
- |
- |
PREDICTED: uncharacterized protein LOC105648579 isoform X1 [Jatropha curcas] |
| 18 |
Hb_004137_020 |
0.1209528796 |
- |
- |
PREDICTED: putative UDP-glucuronate:xylan alpha-glucuronosyltransferase 3 isoform X2 [Jatropha curcas] |
| 19 |
Hb_003746_010 |
0.1231527284 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_032202_040 |
0.1237130684 |
- |
- |
PREDICTED: phospholipase D beta 2 [Jatropha curcas] |