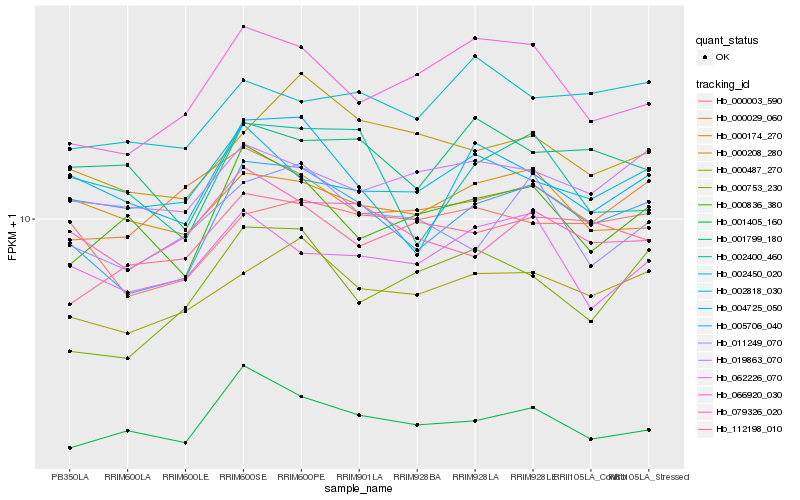
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005706_040 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105642418 [Jatropha curcas] |
| 2 |
Hb_000487_270 |
0.0804052062 |
- |
- |
hypothetical protein JCGZ_22651 [Jatropha curcas] |
| 3 |
Hb_000003_590 |
0.0966806606 |
- |
- |
PREDICTED: uncharacterized protein LOC105631550 isoform X2 [Jatropha curcas] |
| 4 |
Hb_004725_050 |
0.09842807 |
- |
- |
Rhomboid protein, putative [Ricinus communis] |
| 5 |
Hb_066920_030 |
0.1010874682 |
- |
- |
PREDICTED: adagio protein 1 [Jatropha curcas] |
| 6 |
Hb_011249_070 |
0.1016781384 |
- |
- |
hypothetical protein POPTR_0018s12610g [Populus trichocarpa] |
| 7 |
Hb_079326_020 |
0.1022859502 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_000208_280 |
0.1039373162 |
transcription factor |
TF Family: NAC |
PREDICTED: NAC domain-containing protein 78 [Jatropha curcas] |
| 9 |
Hb_019863_070 |
0.1057180475 |
- |
- |
PREDICTED: regulatory-associated protein of TOR 1 isoform X1 [Jatropha curcas] |
| 10 |
Hb_002400_460 |
0.1066556462 |
- |
- |
PREDICTED: uncharacterized protein LOC105637599 isoform X2 [Jatropha curcas] |
| 11 |
Hb_002818_030 |
0.1078636656 |
- |
- |
PREDICTED: protein transport protein SEC23 [Jatropha curcas] |
| 12 |
Hb_000836_380 |
0.1089960111 |
- |
- |
PREDICTED: C-terminal binding protein AN-like [Populus euphratica] |
| 13 |
Hb_112198_010 |
0.1094410775 |
- |
- |
PREDICTED: F-box protein SKIP22 [Jatropha curcas] |
| 14 |
Hb_062226_070 |
0.1097146244 |
transcription factor |
TF Family: FAR1 |
PREDICTED: protein FAR1-RELATED SEQUENCE 4 isoform X1 [Jatropha curcas] |
| 15 |
Hb_000029_060 |
0.1097203657 |
- |
- |
PREDICTED: mRNA-decapping enzyme-like protein [Jatropha curcas] |
| 16 |
Hb_000174_270 |
0.1099255737 |
- |
- |
PREDICTED: type II inositol 1,4,5-trisphosphate 5-phosphatase FRA3 isoform X1 [Jatropha curcas] |
| 17 |
Hb_002450_020 |
0.11020221 |
- |
- |
PREDICTED: probable transcriptional regulator SLK2 [Jatropha curcas] |
| 18 |
Hb_001799_180 |
0.1102046743 |
- |
- |
PREDICTED: serine/threonine-protein kinase D6PK [Jatropha curcas] |
| 19 |
Hb_001405_160 |
0.1106034722 |
- |
- |
PREDICTED: putative pentatricopeptide repeat-containing protein At1g09680 [Jatropha curcas] |
| 20 |
Hb_000753_230 |
0.1109773005 |
- |
- |
PREDICTED: probable galacturonosyltransferase 11 isoform X1 [Jatropha curcas] |