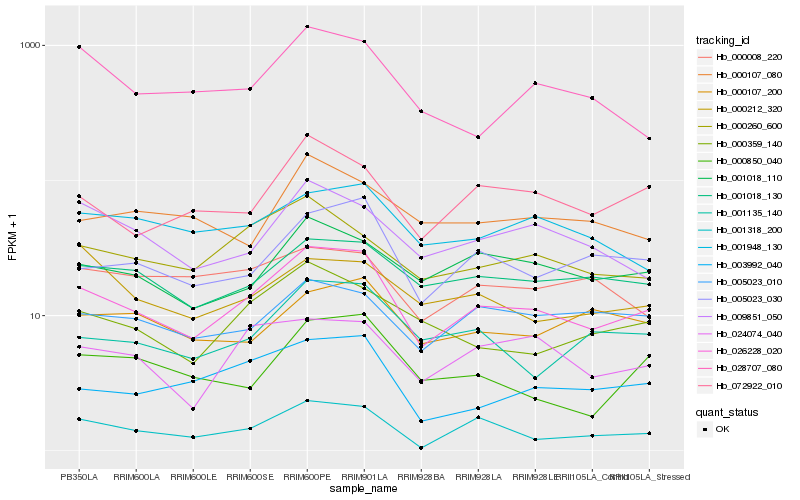
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_026228_020 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105632747 [Jatropha curcas] |
| 2 |
Hb_005023_030 |
0.1160374031 |
- |
- |
PREDICTED: uncharacterized protein LOC103417254 [Malus domestica] |
| 3 |
Hb_072922_010 |
0.1257313874 |
- |
- |
apolipoprotein d, putative [Ricinus communis] |
| 4 |
Hb_001018_130 |
0.1263529881 |
- |
- |
PREDICTED: uncharacterized protein LOC105638260 [Jatropha curcas] |
| 5 |
Hb_001018_110 |
0.1276159284 |
- |
- |
unnamed protein product [Coffea canephora] |
| 6 |
Hb_001318_200 |
0.1294512773 |
- |
- |
PREDICTED: uncharacterized protein LOC105640496 [Jatropha curcas] |
| 7 |
Hb_001135_140 |
0.1295622244 |
- |
- |
PREDICTED: serine/threonine-protein kinase PBS1-like [Malus domestica] |
| 8 |
Hb_003992_040 |
0.131548586 |
- |
- |
Post-illumination chlorophyll fluorescence increase isoform 5 [Theobroma cacao] |
| 9 |
Hb_000260_600 |
0.1315910021 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_000359_140 |
0.1316326312 |
- |
- |
PREDICTED: transmembrane protein 56-like [Jatropha curcas] |
| 11 |
Hb_024074_040 |
0.1330764818 |
- |
- |
PREDICTED: leucine--tRNA ligase, cytoplasmic [Jatropha curcas] |
| 12 |
Hb_009851_050 |
0.1361351507 |
- |
- |
PREDICTED: nephrocystin-3 [Jatropha curcas] |
| 13 |
Hb_005023_010 |
0.1383121776 |
- |
- |
PREDICTED: putative lipase ROG1 [Jatropha curcas] |
| 14 |
Hb_001948_130 |
0.1426715272 |
- |
- |
PREDICTED: uncharacterized protein LOC105638579 [Jatropha curcas] |
| 15 |
Hb_028707_080 |
0.1460523657 |
- |
- |
plasma membrane aquaporin 1 [Hevea brasiliensis] |
| 16 |
Hb_000107_200 |
0.148632603 |
- |
- |
hypothetical protein B456_005G214800 [Gossypium raimondii] |
| 17 |
Hb_000850_040 |
0.1490023002 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_000107_080 |
0.1548924304 |
- |
- |
PREDICTED: uncharacterized protein LOC105641009 [Jatropha curcas] |
| 19 |
Hb_000212_320 |
0.1556512232 |
- |
- |
PREDICTED: uncharacterized protein LOC105644891 [Jatropha curcas] |
| 20 |
Hb_000008_220 |
0.1558687478 |
transcription factor |
TF Family: ARF |
PREDICTED: auxin response factor 10 [Jatropha curcas] |