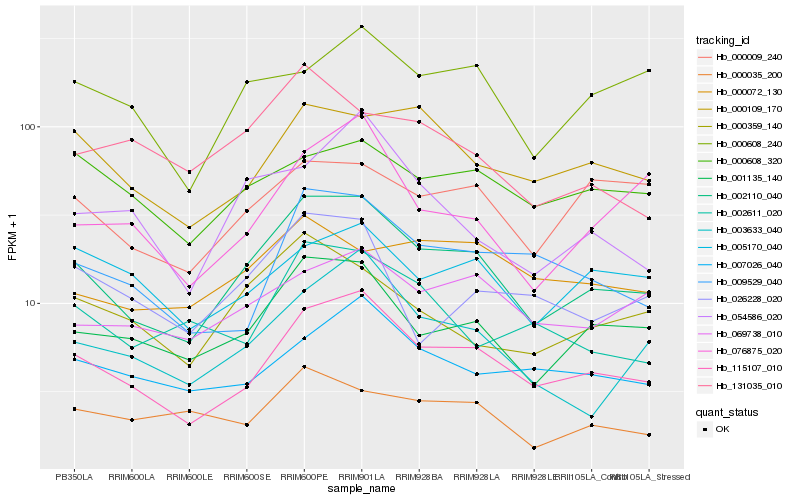
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002110_040 |
0.0 |
- |
- |
hypothetical protein AMTR_s00135p00074190 [Amborella trichopoda] |
| 2 |
Hb_115107_010 |
0.0801516327 |
- |
- |
Early-responsive to dehydration stress protein (ERD4) isoform 1 [Theobroma cacao] |
| 3 |
Hb_001135_140 |
0.1056928396 |
- |
- |
PREDICTED: serine/threonine-protein kinase PBS1-like [Malus domestica] |
| 4 |
Hb_003633_040 |
0.1377092998 |
- |
- |
hypothetical protein B456_008G146200 [Gossypium raimondii] |
| 5 |
Hb_000359_140 |
0.1402182843 |
- |
- |
PREDICTED: transmembrane protein 56-like [Jatropha curcas] |
| 6 |
Hb_000109_170 |
0.1445832753 |
- |
- |
PREDICTED: uncharacterized protein LOC105649868 isoform X1 [Jatropha curcas] |
| 7 |
Hb_009529_040 |
0.1446208496 |
- |
- |
PREDICTED: transmembrane protein 120 homolog isoform X1 [Malus domestica] |
| 8 |
Hb_069738_010 |
0.1469612397 |
- |
- |
PREDICTED: extra-large guanine nucleotide-binding protein 1 [Jatropha curcas] |
| 9 |
Hb_000009_240 |
0.1509229031 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RNF185-like isoform X1 [Jatropha curcas] |
| 10 |
Hb_007026_040 |
0.1539378579 |
- |
- |
PREDICTED: chaperonin 60 subunit alpha 2, chloroplastic-like [Eucalyptus grandis] |
| 11 |
Hb_000035_200 |
0.1547393231 |
- |
- |
PREDICTED: phospholipase D Z-like [Jatropha curcas] |
| 12 |
Hb_000608_240 |
0.1548463407 |
- |
- |
eIF4-gamma/eIF5/eIF2-epsilon domain-containing family protein [Populus trichocarpa] |
| 13 |
Hb_026228_020 |
0.1560658658 |
- |
- |
PREDICTED: uncharacterized protein LOC105632747 [Jatropha curcas] |
| 14 |
Hb_076875_020 |
0.1568412566 |
- |
- |
PREDICTED: alpha/beta hydrolase domain-containing protein 17B [Jatropha curcas] |
| 15 |
Hb_002611_020 |
0.1577739038 |
- |
- |
PREDICTED: alpha-1,3-mannosyl-glycoprotein 2-beta-N-acetylglucosaminyltransferase-like [Populus euphratica] |
| 16 |
Hb_054586_020 |
0.1582322243 |
- |
- |
PREDICTED: exocyst complex component EXO70A1 [Jatropha curcas] |
| 17 |
Hb_000608_320 |
0.158326596 |
- |
- |
PREDICTED: uncharacterized protein LOC105644891 [Jatropha curcas] |
| 18 |
Hb_000072_130 |
0.1585461733 |
- |
- |
hypothetical protein OsJ_28567 [Oryza sativa Japonica Group] |
| 19 |
Hb_005170_040 |
0.1594710278 |
- |
- |
PREDICTED: probable protein S-acyltransferase 7 [Jatropha curcas] |
| 20 |
Hb_131035_010 |
0.1605047546 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |