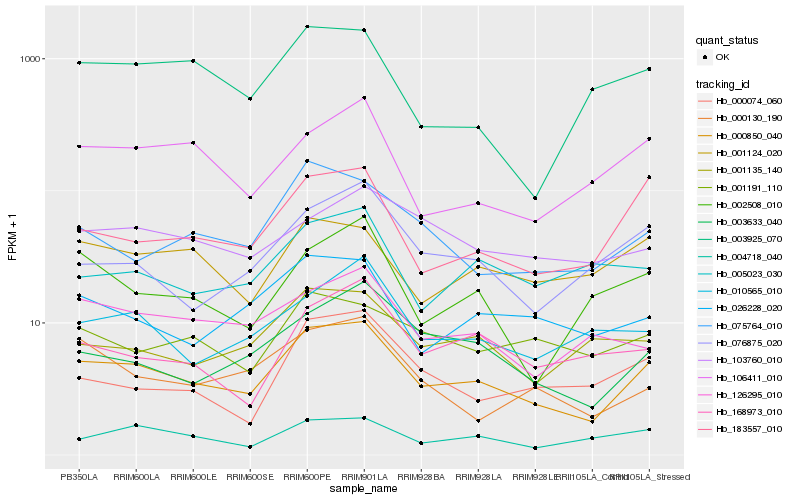
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000850_040 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_183557_010 |
0.1407319592 |
- |
- |
hypothetical protein JCGZ_24847 [Jatropha curcas] |
| 3 |
Hb_003633_040 |
0.1469687678 |
- |
- |
hypothetical protein B456_008G146200 [Gossypium raimondii] |
| 4 |
Hb_075764_010 |
0.1478290708 |
- |
- |
PREDICTED: PRA1 family protein A1-like [Jatropha curcas] |
| 5 |
Hb_026228_020 |
0.1490023002 |
- |
- |
PREDICTED: uncharacterized protein LOC105632747 [Jatropha curcas] |
| 6 |
Hb_004718_040 |
0.1509000352 |
- |
- |
PREDICTED: uncharacterized protein LOC105638292 [Jatropha curcas] |
| 7 |
Hb_106411_010 |
0.1547326759 |
- |
- |
Ribosomal protein S25 family protein [Theobroma cacao] |
| 8 |
Hb_001124_020 |
0.1551462888 |
- |
- |
PREDICTED: protein yippee-like At4g27745 [Jatropha curcas] |
| 9 |
Hb_076875_020 |
0.1552123081 |
- |
- |
PREDICTED: alpha/beta hydrolase domain-containing protein 17B [Jatropha curcas] |
| 10 |
Hb_001135_140 |
0.1553630847 |
- |
- |
PREDICTED: serine/threonine-protein kinase PBS1-like [Malus domestica] |
| 11 |
Hb_126295_010 |
0.1560202311 |
- |
- |
PREDICTED: protein SEH1 [Jatropha curcas] |
| 12 |
Hb_010565_010 |
0.1567108121 |
- |
- |
PREDICTED: N-acetylglucosaminyl-phosphatidylinositol de-N-acetylase-like isoform X2 [Populus euphratica] |
| 13 |
Hb_002508_010 |
0.1579987539 |
- |
- |
snf1-kinase beta subunit, plants, putative [Ricinus communis] |
| 14 |
Hb_000130_190 |
0.159586231 |
- |
- |
PREDICTED: uncharacterized protein LOC105638437 [Jatropha curcas] |
| 15 |
Hb_001191_110 |
0.1610431881 |
- |
- |
sur2 hydroxylase/desaturase, putative [Ricinus communis] |
| 16 |
Hb_168973_010 |
0.1616610522 |
- |
- |
- |
| 17 |
Hb_000074_060 |
0.1632221935 |
- |
- |
PREDICTED: membrane-associated progesterone-binding protein 4-like [Populus euphratica] |
| 18 |
Hb_003925_070 |
0.165548108 |
- |
- |
ATOZI1, putative [Ricinus communis] |
| 19 |
Hb_005023_030 |
0.165883042 |
- |
- |
PREDICTED: uncharacterized protein LOC103417254 [Malus domestica] |
| 20 |
Hb_103760_010 |
0.1680106926 |
- |
- |
serine/arginine rich splicing factor, putative [Ricinus communis] |