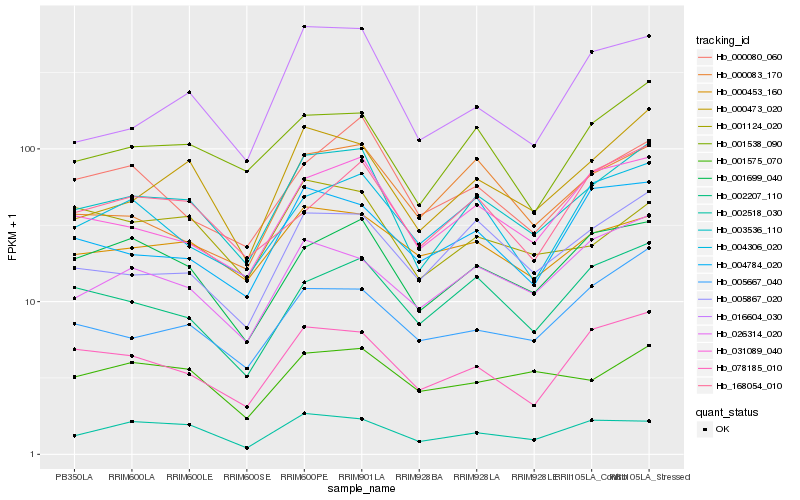
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003536_110 |
0.0 |
- |
- |
PREDICTED: probable small nuclear ribonucleoprotein F [Jatropha curcas] |
| 2 |
Hb_001538_090 |
0.0956427326 |
- |
- |
40S ribosomal protein S29 [Morus notabilis] |
| 3 |
Hb_000473_020 |
0.0981682449 |
- |
- |
expressed protein, putative [Ricinus communis] |
| 4 |
Hb_031089_040 |
0.0986209304 |
- |
- |
thioredoxin H-type 1 [Hevea brasiliensis] |
| 5 |
Hb_078185_010 |
0.1040518157 |
- |
- |
- |
| 6 |
Hb_001699_040 |
0.1044765322 |
- |
- |
PREDICTED: uncharacterized protein LOC105636529 isoform X2 [Jatropha curcas] |
| 7 |
Hb_005867_020 |
0.1091281944 |
- |
- |
PREDICTED: sm-like protein LSM8 [Jatropha curcas] |
| 8 |
Hb_026314_020 |
0.1148992125 |
- |
- |
hypothetical protein POPTR_0019s10050g [Populus trichocarpa] |
| 9 |
Hb_004306_020 |
0.1176271734 |
- |
- |
PREDICTED: ankyrin repeat and SAM domain-containing protein 6 [Jatropha curcas] |
| 10 |
Hb_000080_060 |
0.1187489502 |
- |
- |
RNA polymerase I, II and III subunit RPB8, partial [Manihot esculenta] |
| 11 |
Hb_002207_110 |
0.1188759355 |
- |
- |
- |
| 12 |
Hb_002518_030 |
0.1194754826 |
- |
- |
PREDICTED: phosphatidylinositol:ceramide inositolphosphotransferase 3 isoform X1 [Populus euphratica] |
| 13 |
Hb_000453_160 |
0.1208410461 |
- |
- |
vacuolar protein sorting 26, vps26, putative [Ricinus communis] |
| 14 |
Hb_005667_040 |
0.1213554782 |
- |
- |
PREDICTED: trafficking protein particle complex subunit 5 [Jatropha curcas] |
| 15 |
Hb_004784_020 |
0.1235675316 |
- |
- |
PREDICTED: uncharacterized protein LOC105636452 [Jatropha curcas] |
| 16 |
Hb_001124_020 |
0.1255006983 |
- |
- |
PREDICTED: protein yippee-like At4g27745 [Jatropha curcas] |
| 17 |
Hb_168054_010 |
0.1261804499 |
- |
- |
PREDICTED: peptidyl-prolyl cis-trans isomerase CYP18-1 [Elaeis guineensis] |
| 18 |
Hb_000083_170 |
0.1277683469 |
- |
- |
PREDICTED: DNA-directed RNA polymerase V subunit 7-like [Populus euphratica] |
| 19 |
Hb_016604_030 |
0.1279422588 |
- |
- |
histone H4 [Zea mays] |
| 20 |
Hb_001575_070 |
0.128158708 |
- |
- |
PREDICTED: uncharacterized protein LOC105639885 [Jatropha curcas] |