| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
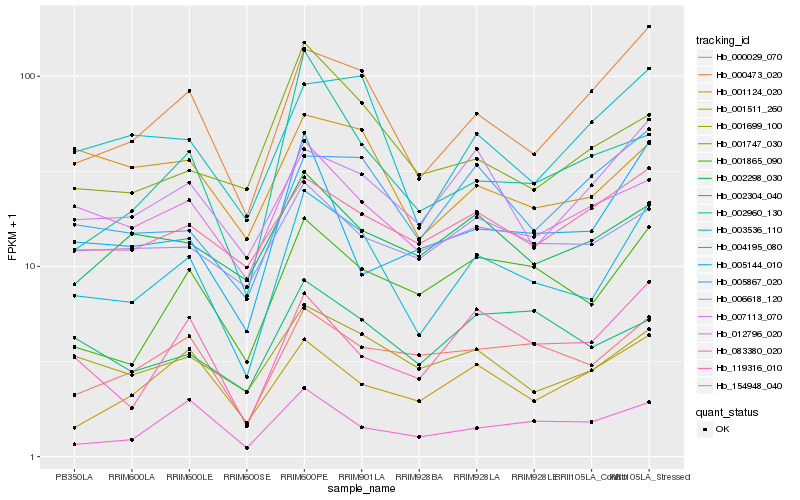
Hb_004195_080 |
0.0 |
- |
- |
PREDICTED: cyclin-P3-1 isoform X2 [Jatropha curcas] |
| 2 |
Hb_000473_020 |
0.112569552 |
- |
- |
expressed protein, putative [Ricinus communis] |
| 3 |
Hb_003536_110 |
0.1348907014 |
- |
- |
PREDICTED: probable small nuclear ribonucleoprotein F [Jatropha curcas] |
| 4 |
Hb_001865_090 |
0.1369269853 |
- |
- |
hypothetical protein VITISV_005773 [Vitis vinifera] |
| 5 |
Hb_000029_070 |
0.1431064766 |
- |
- |
PREDICTED: cold-inducible RNA-binding protein B-like [Jatropha curcas] |
| 6 |
Hb_012796_020 |
0.145187003 |
- |
- |
PREDICTED: ADP-ribosylation factor 1 [Prunus mume] |
| 7 |
Hb_005867_020 |
0.1474685714 |
- |
- |
PREDICTED: sm-like protein LSM8 [Jatropha curcas] |
| 8 |
Hb_001699_100 |
0.1491371779 |
- |
- |
PREDICTED: calmodulin-lysine N-methyltransferase [Jatropha curcas] |
| 9 |
Hb_119316_010 |
0.1498166586 |
- |
- |
tryptophan biosynthesis protein, putative [Ricinus communis] |
| 10 |
Hb_007113_070 |
0.1500571486 |
- |
- |
protein with unknown function [Ricinus communis] |
| 11 |
Hb_001124_020 |
0.1569414734 |
- |
- |
PREDICTED: protein yippee-like At4g27745 [Jatropha curcas] |
| 12 |
Hb_154948_040 |
0.1575413852 |
- |
- |
hypothetical protein B456_013G043800 [Gossypium raimondii] |
| 13 |
Hb_002960_130 |
0.1594560672 |
- |
- |
hypothetical protein B456_008G036100 [Gossypium raimondii] |
| 14 |
Hb_002304_040 |
0.1607309975 |
- |
- |
PREDICTED: uncharacterized protein LOC105649623 isoform X2 [Jatropha curcas] |
| 15 |
Hb_002298_030 |
0.1616202769 |
- |
- |
PREDICTED: coenzyme Q-binding protein COQ10 homolog, mitochondrial [Jatropha curcas] |
| 16 |
Hb_001747_030 |
0.1616875434 |
- |
- |
hypothetical protein F383_08210 [Gossypium arboreum] |
| 17 |
Hb_006618_120 |
0.1618594309 |
- |
- |
PREDICTED: uncharacterized protein LOC105645969 [Jatropha curcas] |
| 18 |
Hb_083380_020 |
0.1629795237 |
- |
- |
PREDICTED: LAG1 longevity assurance homolog 2 [Jatropha curcas] |
| 19 |
Hb_001511_260 |
0.1634569703 |
- |
- |
tryptophan biosynthesis protein, putative [Ricinus communis] |
| 20 |
Hb_005144_010 |
0.1651864937 |
- |
- |
PREDICTED: lactoylglutathione lyase [Jatropha curcas] |