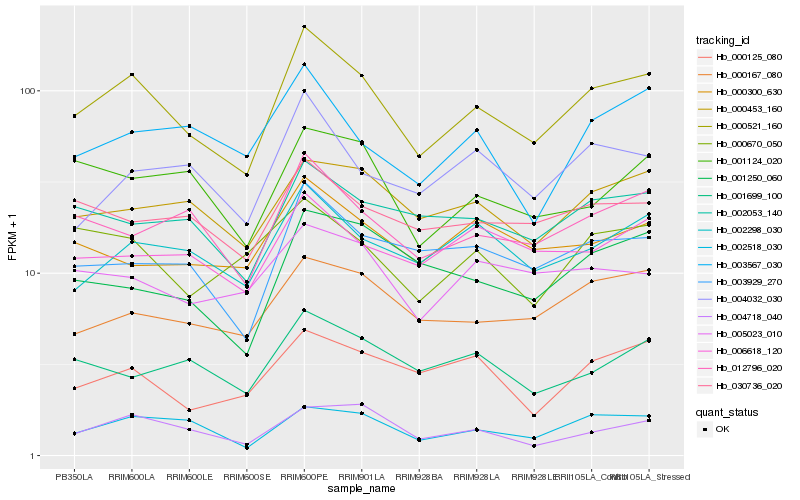
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000521_160 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_002518_030 |
0.1112150018 |
- |
- |
PREDICTED: phosphatidylinositol:ceramide inositolphosphotransferase 3 isoform X1 [Populus euphratica] |
| 3 |
Hb_012796_020 |
0.1113308757 |
- |
- |
PREDICTED: ADP-ribosylation factor 1 [Prunus mume] |
| 4 |
Hb_002298_030 |
0.1146743797 |
- |
- |
PREDICTED: coenzyme Q-binding protein COQ10 homolog, mitochondrial [Jatropha curcas] |
| 5 |
Hb_003929_270 |
0.114709935 |
- |
- |
S-adenosylmethionine-dependent methyltransferase, putative [Ricinus communis] |
| 6 |
Hb_000167_080 |
0.1178664098 |
- |
- |
PREDICTED: uncharacterized protein LOC105641103 [Jatropha curcas] |
| 7 |
Hb_000453_160 |
0.1197843205 |
- |
- |
vacuolar protein sorting 26, vps26, putative [Ricinus communis] |
| 8 |
Hb_004718_040 |
0.1204965378 |
- |
- |
PREDICTED: uncharacterized protein LOC105638292 [Jatropha curcas] |
| 9 |
Hb_002053_140 |
0.1210616814 |
- |
- |
PREDICTED: coatomer subunit epsilon-1 [Jatropha curcas] |
| 10 |
Hb_030736_020 |
0.126622239 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_001124_020 |
0.1270046001 |
- |
- |
PREDICTED: protein yippee-like At4g27745 [Jatropha curcas] |
| 12 |
Hb_000670_050 |
0.1275154455 |
transcription factor |
TF Family: MYB |
hypothetical protein JCGZ_16088 [Jatropha curcas] |
| 13 |
Hb_006618_120 |
0.1279467912 |
- |
- |
PREDICTED: uncharacterized protein LOC105645969 [Jatropha curcas] |
| 14 |
Hb_004032_030 |
0.1281863677 |
- |
- |
PREDICTED: RNA-binding protein 24-like isoform X1 [Populus euphratica] |
| 15 |
Hb_000125_080 |
0.1283718903 |
transcription factor |
TF Family: mTERF |
- |
| 16 |
Hb_001699_100 |
0.1294718533 |
- |
- |
PREDICTED: calmodulin-lysine N-methyltransferase [Jatropha curcas] |
| 17 |
Hb_001250_060 |
0.1300386431 |
- |
- |
PREDICTED: signal recognition particle receptor subunit beta-like [Jatropha curcas] |
| 18 |
Hb_003567_030 |
0.1306301147 |
- |
- |
PREDICTED: nifU-like protein 4, mitochondrial [Jatropha curcas] |
| 19 |
Hb_005023_010 |
0.1307144576 |
- |
- |
PREDICTED: putative lipase ROG1 [Jatropha curcas] |
| 20 |
Hb_000300_630 |
0.1307282209 |
- |
- |
PREDICTED: 5'-adenylylsulfate reductase-like 7 [Jatropha curcas] |