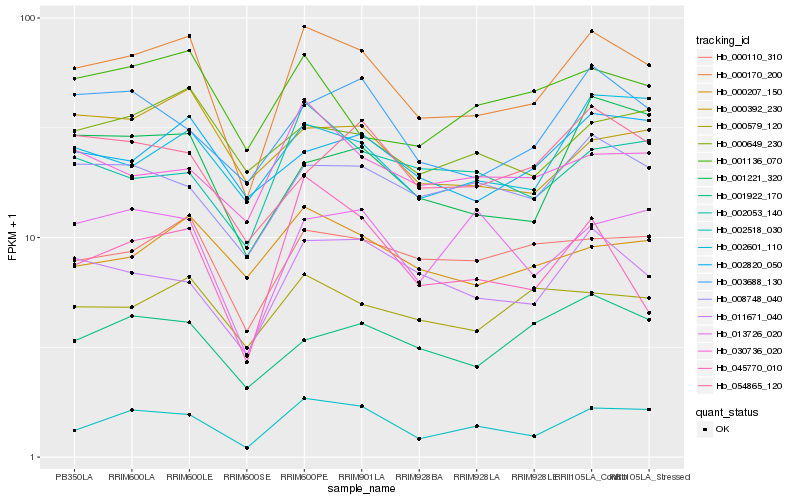
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000170_200 |
0.0 |
- |
- |
PREDICTED: protein KRTCAP2 homolog [Jatropha curcas] |
| 2 |
Hb_002518_030 |
0.0922101587 |
- |
- |
PREDICTED: phosphatidylinositol:ceramide inositolphosphotransferase 3 isoform X1 [Populus euphratica] |
| 3 |
Hb_011671_040 |
0.1011086589 |
- |
- |
PREDICTED: uncharacterized protein LOC105646469 isoform X2 [Jatropha curcas] |
| 4 |
Hb_000110_310 |
0.103572786 |
rubber biosynthesis |
Gene Name: Dihydrolipoyllysine-residue acetyltransferase component 4 of pyruvate dehydrogenase complex |
Dihydrolipoyllysine-residue acetyltransferase component of pyruvate dehydrogenase, putative [Ricinus communis] |
| 5 |
Hb_008748_040 |
0.1080620882 |
- |
- |
beta-hydroxyacyl-acyl carrier protein dehydratase [Hevea brasiliensis] |
| 6 |
Hb_003688_130 |
0.1121954081 |
- |
- |
phosphoserine phosphatase, putative [Ricinus communis] |
| 7 |
Hb_000579_120 |
0.1126585184 |
- |
- |
PREDICTED: uncharacterized protein LOC105633845 [Jatropha curcas] |
| 8 |
Hb_002820_050 |
0.1142728404 |
- |
- |
PREDICTED: putative glutathione-specific gamma-glutamylcyclotransferase 2 [Jatropha curcas] |
| 9 |
Hb_000207_150 |
0.1167280704 |
- |
- |
PREDICTED: uncharacterized membrane protein At4g09580 [Jatropha curcas] |
| 10 |
Hb_002601_110 |
0.1169421827 |
- |
- |
PREDICTED: probable histone H2A variant 3 [Jatropha curcas] |
| 11 |
Hb_001221_320 |
0.1173020442 |
transcription factor |
TF Family: NF-YC |
PREDICTED: nuclear transcription factor Y subunit C-1 isoform X1 [Jatropha curcas] |
| 12 |
Hb_001922_170 |
0.117969599 |
- |
- |
Plastidic pyruvate kinase beta subunit 1 isoform 2 [Theobroma cacao] |
| 13 |
Hb_000649_230 |
0.1194408722 |
- |
- |
PREDICTED: ubiquitin-conjugating enzyme E2 variant 1A-like [Jatropha curcas] |
| 14 |
Hb_000392_230 |
0.1198540909 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_054865_120 |
0.12011675 |
- |
- |
PREDICTED: CBL-interacting serine/threonine-protein kinase 9 [Jatropha curcas] |
| 16 |
Hb_045770_010 |
0.1202075671 |
- |
- |
ER lumen protein retaining receptor [Populus trichocarpa] |
| 17 |
Hb_002053_140 |
0.120623921 |
- |
- |
PREDICTED: coatomer subunit epsilon-1 [Jatropha curcas] |
| 18 |
Hb_001136_070 |
0.1209412299 |
- |
- |
Transmembrane protein TPARL, putative [Ricinus communis] |
| 19 |
Hb_030736_020 |
0.1217393727 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_013726_020 |
0.1233286263 |
- |
- |
ara4-interacting protein, putative [Ricinus communis] |