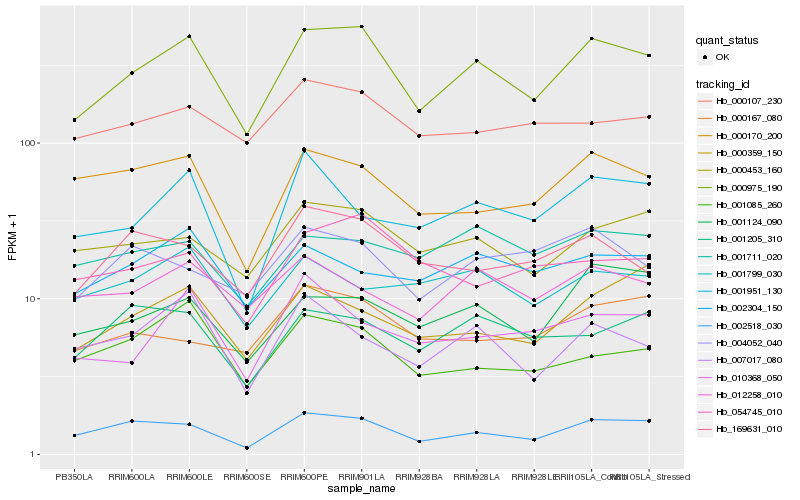
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000975_190 |
0.0 |
- |
- |
histone H4 [Zea mays] |
| 2 |
Hb_002518_030 |
0.1000972181 |
- |
- |
PREDICTED: phosphatidylinositol:ceramide inositolphosphotransferase 3 isoform X1 [Populus euphratica] |
| 3 |
Hb_000453_160 |
0.1171190008 |
- |
- |
vacuolar protein sorting 26, vps26, putative [Ricinus communis] |
| 4 |
Hb_001124_090 |
0.1304345737 |
- |
- |
acyl carrier protein, putative [Ricinus communis] |
| 5 |
Hb_000170_200 |
0.1307880386 |
- |
- |
PREDICTED: protein KRTCAP2 homolog [Jatropha curcas] |
| 6 |
Hb_169631_010 |
0.1309002754 |
- |
- |
MADS-box transcription factor, putative [Ricinus communis] |
| 7 |
Hb_012258_010 |
0.1327378597 |
- |
- |
PREDICTED: cyclase-associated protein 1 [Jatropha curcas] |
| 8 |
Hb_000107_230 |
0.1328725435 |
- |
- |
unknown [Medicago truncatula] |
| 9 |
Hb_002304_150 |
0.1351839736 |
- |
- |
PREDICTED: protein IQ-DOMAIN 1-like isoform X1 [Jatropha curcas] |
| 10 |
Hb_007017_080 |
0.1358525757 |
transcription factor |
TF Family: bHLH |
DNA binding protein, putative [Ricinus communis] |
| 11 |
Hb_054745_010 |
0.1367822215 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_001205_310 |
0.1371462093 |
- |
- |
PREDICTED: TPR repeat-containing thioredoxin TTL4-like [Jatropha curcas] |
| 13 |
Hb_001711_020 |
0.1374804637 |
- |
- |
PREDICTED: AP-2 complex subunit mu [Jatropha curcas] |
| 14 |
Hb_001085_260 |
0.1379203344 |
- |
- |
PREDICTED: uncharacterized protein LOC105633184 [Jatropha curcas] |
| 15 |
Hb_004052_040 |
0.1383615077 |
- |
- |
PREDICTED: putative G3BP-like protein [Jatropha curcas] |
| 16 |
Hb_001951_130 |
0.1393435823 |
- |
- |
PREDICTED: coatomer subunit zeta-1-like [Jatropha curcas] |
| 17 |
Hb_010368_050 |
0.1399148747 |
- |
- |
PREDICTED: uncharacterized protein C1450.15 [Jatropha curcas] |
| 18 |
Hb_001799_030 |
0.1408942015 |
- |
- |
PREDICTED: ELMO domain-containing protein C-like [Jatropha curcas] |
| 19 |
Hb_000167_080 |
0.1409486843 |
- |
- |
PREDICTED: uncharacterized protein LOC105641103 [Jatropha curcas] |
| 20 |
Hb_000359_150 |
0.1417008794 |
- |
- |
PREDICTED: protein Mpv17-like isoform X1 [Jatropha curcas] |