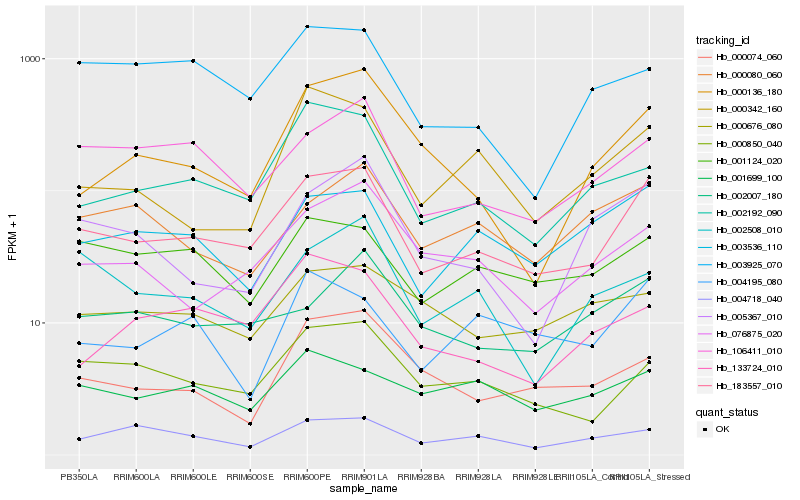
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_183557_010 |
0.0 |
- |
- |
hypothetical protein JCGZ_24847 [Jatropha curcas] |
| 2 |
Hb_000850_040 |
0.1407319592 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_003536_110 |
0.1464909082 |
- |
- |
PREDICTED: probable small nuclear ribonucleoprotein F [Jatropha curcas] |
| 4 |
Hb_106411_010 |
0.1493050941 |
- |
- |
Ribosomal protein S25 family protein [Theobroma cacao] |
| 5 |
Hb_076875_020 |
0.1499652446 |
- |
- |
PREDICTED: alpha/beta hydrolase domain-containing protein 17B [Jatropha curcas] |
| 6 |
Hb_002192_090 |
0.1574128228 |
- |
- |
hypothetical protein AMTR_s00029p00055000 [Amborella trichopoda] |
| 7 |
Hb_001124_020 |
0.1590246945 |
- |
- |
PREDICTED: protein yippee-like At4g27745 [Jatropha curcas] |
| 8 |
Hb_002007_180 |
0.1616099787 |
- |
- |
PREDICTED: uncharacterized protein LOC105647525 [Jatropha curcas] |
| 9 |
Hb_000080_060 |
0.1701658964 |
- |
- |
RNA polymerase I, II and III subunit RPB8, partial [Manihot esculenta] |
| 10 |
Hb_005367_010 |
0.1702604927 |
- |
- |
- |
| 11 |
Hb_004718_040 |
0.1704748517 |
- |
- |
PREDICTED: uncharacterized protein LOC105638292 [Jatropha curcas] |
| 12 |
Hb_000074_060 |
0.171725848 |
- |
- |
PREDICTED: membrane-associated progesterone-binding protein 4-like [Populus euphratica] |
| 13 |
Hb_001699_100 |
0.1732093354 |
- |
- |
PREDICTED: calmodulin-lysine N-methyltransferase [Jatropha curcas] |
| 14 |
Hb_133724_010 |
0.1747368598 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_000676_080 |
0.1749396849 |
- |
- |
PREDICTED: uncharacterized WD repeat-containing protein C17D11.16 [Jatropha curcas] |
| 16 |
Hb_000136_180 |
0.1751216129 |
- |
- |
ribosomal protein S28, putative [Ricinus communis] |
| 17 |
Hb_000342_160 |
0.1757671269 |
- |
- |
PREDICTED: sarcoplasmic reticulum histidine-rich calcium-binding protein [Jatropha curcas] |
| 18 |
Hb_004195_080 |
0.1761917 |
- |
- |
PREDICTED: cyclin-P3-1 isoform X2 [Jatropha curcas] |
| 19 |
Hb_003925_070 |
0.1764200367 |
- |
- |
ATOZI1, putative [Ricinus communis] |
| 20 |
Hb_002508_010 |
0.1767456567 |
- |
- |
snf1-kinase beta subunit, plants, putative [Ricinus communis] |