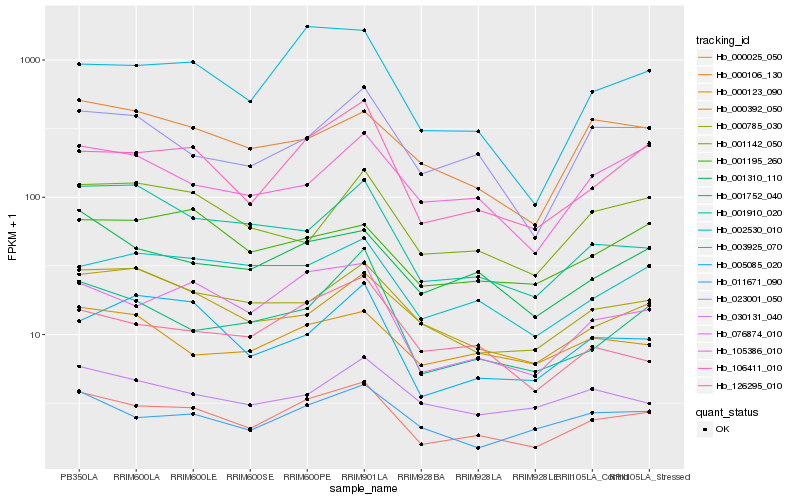
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000025_050 |
0.0 |
- |
- |
hypothetical protein POPTR_0003s13040g [Populus trichocarpa] |
| 2 |
Hb_076874_010 |
0.0800086485 |
- |
- |
PREDICTED: ER lumen protein-retaining receptor [Jatropha curcas] |
| 3 |
Hb_106411_010 |
0.0948082642 |
- |
- |
Ribosomal protein S25 family protein [Theobroma cacao] |
| 4 |
Hb_001310_110 |
0.1016668405 |
- |
- |
unnamed protein product [Coffea canephora] |
| 5 |
Hb_011671_090 |
0.1091233305 |
- |
- |
PREDICTED: protein RFT1 homolog isoform X1 [Jatropha curcas] |
| 6 |
Hb_000785_030 |
0.109494233 |
- |
- |
hypothetical protein RCOM_0808030 [Ricinus communis] |
| 7 |
Hb_003925_070 |
0.1107530756 |
- |
- |
ATOZI1, putative [Ricinus communis] |
| 8 |
Hb_002530_010 |
0.1118119447 |
- |
- |
hypothetical protein EUGRSUZ_C02580 [Eucalyptus grandis] |
| 9 |
Hb_023001_050 |
0.1121307594 |
- |
- |
hypothetical protein POPTR_0006s10330g [Populus trichocarpa] |
| 10 |
Hb_126295_010 |
0.1181843768 |
- |
- |
PREDICTED: protein SEH1 [Jatropha curcas] |
| 11 |
Hb_001752_040 |
0.1205434051 |
- |
- |
hypothetical protein EUGRSUZ_C02580 [Eucalyptus grandis] |
| 12 |
Hb_000123_090 |
0.1206309242 |
transcription factor |
TF Family: B3 |
PREDICTED: B3 domain-containing protein Os03g0620400-like [Jatropha curcas] |
| 13 |
Hb_105386_010 |
0.1226024522 |
- |
- |
PREDICTED: 60S ribosomal protein L7-2-like [Jatropha curcas] |
| 14 |
Hb_000106_130 |
0.1227291648 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 15 |
Hb_001910_020 |
0.1257340741 |
- |
- |
PREDICTED: ubiquitin-associated domain-containing protein 2 [Jatropha curcas] |
| 16 |
Hb_005085_020 |
0.1265691615 |
- |
- |
hypothetical protein B456_001G193200 [Gossypium raimondii] |
| 17 |
Hb_000392_050 |
0.1273807083 |
- |
- |
PREDICTED: uncharacterized protein LOC105640540 isoform X2 [Jatropha curcas] |
| 18 |
Hb_030131_040 |
0.1275946319 |
- |
- |
ribosome biogenesis protein tsr1, putative [Ricinus communis] |
| 19 |
Hb_001195_260 |
0.1276148469 |
- |
- |
Mitochondrial 50S ribosomal protein L27 [Hevea brasiliensis] |
| 20 |
Hb_001142_050 |
0.1283646432 |
- |
- |
PREDICTED: coiled-coil domain-containing protein 12 [Jatropha curcas] |