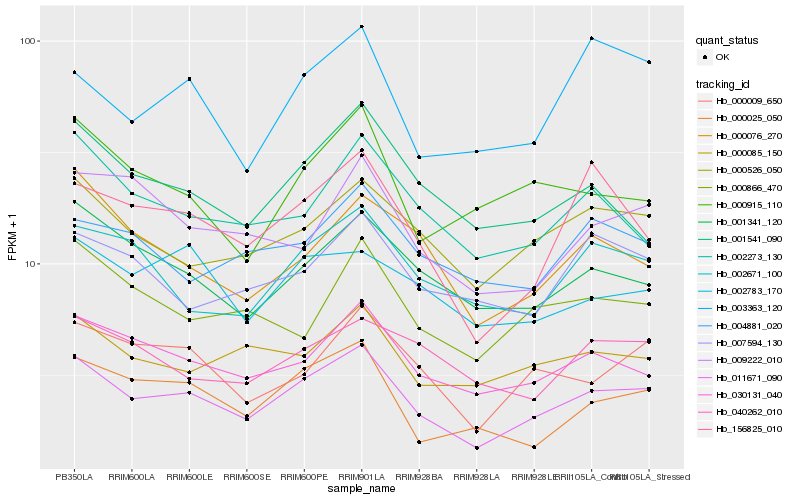
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_011671_090 |
0.0 |
- |
- |
PREDICTED: protein RFT1 homolog isoform X1 [Jatropha curcas] |
| 2 |
Hb_030131_040 |
0.0921795626 |
- |
- |
ribosome biogenesis protein tsr1, putative [Ricinus communis] |
| 3 |
Hb_000526_050 |
0.09850328 |
- |
- |
PREDICTED: chaperone protein dnaJ 1, mitochondrial [Jatropha curcas] |
| 4 |
Hb_001341_120 |
0.1023391733 |
- |
- |
hypothetical protein B456_001G130300 [Gossypium raimondii] |
| 5 |
Hb_000009_650 |
0.1028358474 |
- |
- |
PREDICTED: ribosomal RNA small subunit methyltransferase nep-1 [Jatropha curcas] |
| 6 |
Hb_156825_010 |
0.10817467 |
transcription factor |
TF Family: C2C2-GATA |
hypothetical protein JCGZ_05054 [Jatropha curcas] |
| 7 |
Hb_000025_050 |
0.1091233305 |
- |
- |
hypothetical protein POPTR_0003s13040g [Populus trichocarpa] |
| 8 |
Hb_002671_100 |
0.1099994892 |
- |
- |
PREDICTED: tRNA (guanine-N(7)-)-methyltransferase-like [Jatropha curcas] |
| 9 |
Hb_000085_150 |
0.1100622257 |
- |
- |
PREDICTED: putative BPI/LBP family protein At1g04970 [Jatropha curcas] |
| 10 |
Hb_002273_130 |
0.1120081175 |
- |
- |
PREDICTED: T-complex protein 1 subunit eta [Gossypium raimondii] |
| 11 |
Hb_001541_090 |
0.1131115781 |
- |
- |
PREDICTED: uncharacterized protein At1g04910 isoform X1 [Jatropha curcas] |
| 12 |
Hb_000076_270 |
0.1154212526 |
- |
- |
PREDICTED: DDRGK domain-containing protein 1 [Populus euphratica] |
| 13 |
Hb_002783_170 |
0.1159601188 |
- |
- |
PREDICTED: BRCA1-A complex subunit BRE isoform X2 [Jatropha curcas] |
| 14 |
Hb_000866_470 |
0.117297421 |
- |
- |
PREDICTED: uncharacterized protein LOC105643015 [Jatropha curcas] |
| 15 |
Hb_000915_110 |
0.1178058182 |
- |
- |
PREDICTED: peptide methionine sulfoxide reductase A5 [Jatropha curcas] |
| 16 |
Hb_009222_010 |
0.1195210218 |
- |
- |
WD-repeat protein, putative [Ricinus communis] |
| 17 |
Hb_007594_130 |
0.11954655 |
- |
- |
PREDICTED: lysophospholipid acyltransferase LPEAT2 isoform X1 [Jatropha curcas] |
| 18 |
Hb_040262_010 |
0.1200660514 |
- |
- |
PREDICTED: aspartate carbamoyltransferase 1, chloroplastic [Jatropha curcas] |
| 19 |
Hb_004881_020 |
0.1201195532 |
- |
- |
PREDICTED: serine/threonine-protein kinase rio2-like [Jatropha curcas] |
| 20 |
Hb_003363_120 |
0.1217874597 |
- |
- |
GTP-binding family protein [Populus trichocarpa] |