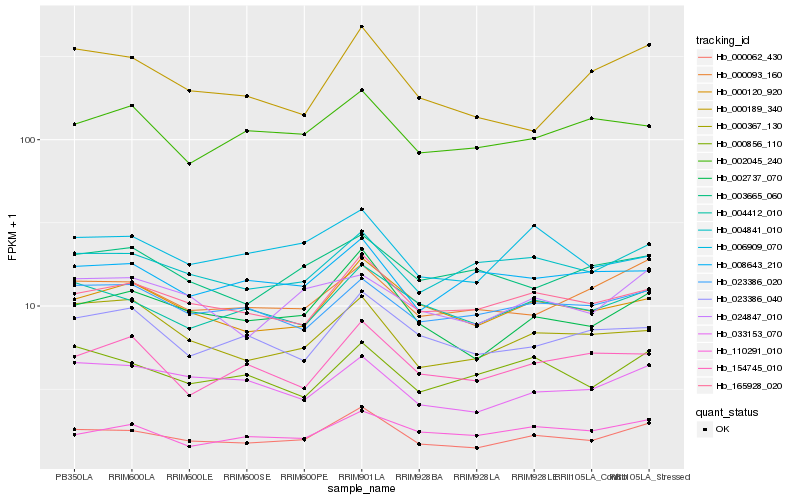
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000062_430 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105644764 isoform X1 [Jatropha curcas] |
| 2 |
Hb_002737_070 |
0.0561051146 |
- |
- |
PREDICTED: phagocyte signaling-impaired protein [Jatropha curcas] |
| 3 |
Hb_165928_020 |
0.0700106285 |
- |
- |
PREDICTED: zinc finger protein VAR3, chloroplastic [Jatropha curcas] |
| 4 |
Hb_000120_920 |
0.0733786619 |
- |
- |
poly(A) polymerase, putative [Ricinus communis] |
| 5 |
Hb_004412_010 |
0.0763979881 |
- |
- |
- |
| 6 |
Hb_004841_010 |
0.0843063682 |
- |
- |
PREDICTED: 60S ribosomal protein L2, mitochondrial [Jatropha curcas] |
| 7 |
Hb_000093_160 |
0.0866838323 |
- |
- |
PREDICTED: nuclear transcription factor Y subunit B-1-like isoform X1 [Jatropha curcas] |
| 8 |
Hb_033153_070 |
0.0920309959 |
- |
- |
PREDICTED: uncharacterized protein LOC105649606 isoform X1 [Jatropha curcas] |
| 9 |
Hb_110291_010 |
0.0927655653 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_023386_040 |
0.0939544126 |
- |
- |
PREDICTED: chaperone protein dnaJ 13-like [Populus euphratica] |
| 11 |
Hb_023386_020 |
0.0950000068 |
- |
- |
PREDICTED: chaperone protein dnaJ 13 isoform X2 [Jatropha curcas] |
| 12 |
Hb_006909_070 |
0.0974883874 |
- |
- |
ABC transporter family protein [Hevea brasiliensis] |
| 13 |
Hb_000856_110 |
0.0975205945 |
- |
- |
PREDICTED: uncharacterized protein LOC105640477 isoform X2 [Jatropha curcas] |
| 14 |
Hb_002045_240 |
0.0976297765 |
- |
- |
2-methyl-6-phytylbenzoquinone methyltranferase [Hevea brasiliensis] |
| 15 |
Hb_154745_010 |
0.0982482161 |
- |
- |
dead box ATP-dependent RNA helicase, putative [Ricinus communis] |
| 16 |
Hb_024847_010 |
0.0985672359 |
- |
- |
UPF0586 C9orf41-like protein [Medicago truncatula] |
| 17 |
Hb_000367_130 |
0.099151082 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_008643_210 |
0.0995977689 |
- |
- |
PREDICTED: uncharacterized protein LOC105644223 isoform X1 [Jatropha curcas] |
| 19 |
Hb_003665_060 |
0.1009541958 |
- |
- |
PREDICTED: protein N-lysine methyltransferase METTL21A [Jatropha curcas] |
| 20 |
Hb_000189_340 |
0.1012792817 |
- |
- |
PREDICTED: 40S ribosomal protein S3a-1 [Jatropha curcas] |