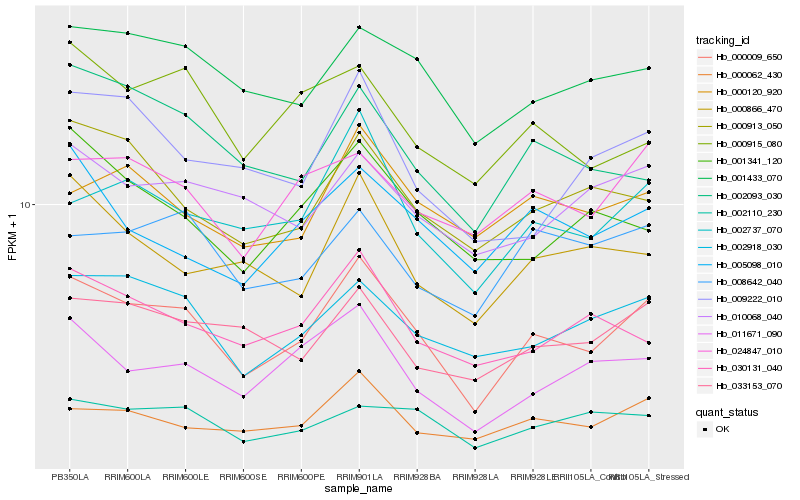
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000009_650 |
0.0 |
- |
- |
PREDICTED: ribosomal RNA small subunit methyltransferase nep-1 [Jatropha curcas] |
| 2 |
Hb_002737_070 |
0.10012173 |
- |
- |
PREDICTED: phagocyte signaling-impaired protein [Jatropha curcas] |
| 3 |
Hb_002918_030 |
0.100584461 |
- |
- |
PREDICTED: dol-P-Glc:Glc(2)Man(9)GlcNAc(2)-PP-Dol alpha-1,2-glucosyltransferase isoform X1 [Jatropha curcas] |
| 4 |
Hb_011671_090 |
0.1028358474 |
- |
- |
PREDICTED: protein RFT1 homolog isoform X1 [Jatropha curcas] |
| 5 |
Hb_000062_430 |
0.1056989282 |
- |
- |
PREDICTED: uncharacterized protein LOC105644764 isoform X1 [Jatropha curcas] |
| 6 |
Hb_001433_070 |
0.1075622739 |
- |
- |
PREDICTED: splicing regulatory glutamine/lysine-rich protein 1 [Jatropha curcas] |
| 7 |
Hb_000913_050 |
0.1109804913 |
- |
- |
hypothetical protein JCGZ_17329 [Jatropha curcas] |
| 8 |
Hb_024847_010 |
0.1122300316 |
- |
- |
UPF0586 C9orf41-like protein [Medicago truncatula] |
| 9 |
Hb_002110_230 |
0.1130555479 |
- |
- |
PREDICTED: protein TRANSPARENT TESTA 12-like [Jatropha curcas] |
| 10 |
Hb_033153_070 |
0.1132514376 |
- |
- |
PREDICTED: uncharacterized protein LOC105649606 isoform X1 [Jatropha curcas] |
| 11 |
Hb_009222_010 |
0.1133243918 |
- |
- |
WD-repeat protein, putative [Ricinus communis] |
| 12 |
Hb_000866_470 |
0.1162684252 |
- |
- |
PREDICTED: uncharacterized protein LOC105643015 [Jatropha curcas] |
| 13 |
Hb_030131_040 |
0.116329147 |
- |
- |
ribosome biogenesis protein tsr1, putative [Ricinus communis] |
| 14 |
Hb_002093_030 |
0.1169523625 |
- |
- |
PREDICTED: uncharacterized protein LOC105648523 [Jatropha curcas] |
| 15 |
Hb_005098_010 |
0.118400738 |
- |
- |
PREDICTED: L-aminoadipate-semialdehyde dehydrogenase-phosphopantetheinyl transferase-like [Jatropha curcas] |
| 16 |
Hb_001341_120 |
0.1186396783 |
- |
- |
hypothetical protein B456_001G130300 [Gossypium raimondii] |
| 17 |
Hb_000120_920 |
0.1187928878 |
- |
- |
poly(A) polymerase, putative [Ricinus communis] |
| 18 |
Hb_008642_040 |
0.1189262221 |
- |
- |
PREDICTED: nucleolar GTP-binding protein 1 isoform X2 [Jatropha curcas] |
| 19 |
Hb_000915_080 |
0.1193984835 |
- |
- |
PREDICTED: uncharacterized protein LOC105628777 [Jatropha curcas] |
| 20 |
Hb_010068_040 |
0.1196809345 |
- |
- |
PREDICTED: uncharacterized protein LOC105632964 [Jatropha curcas] |