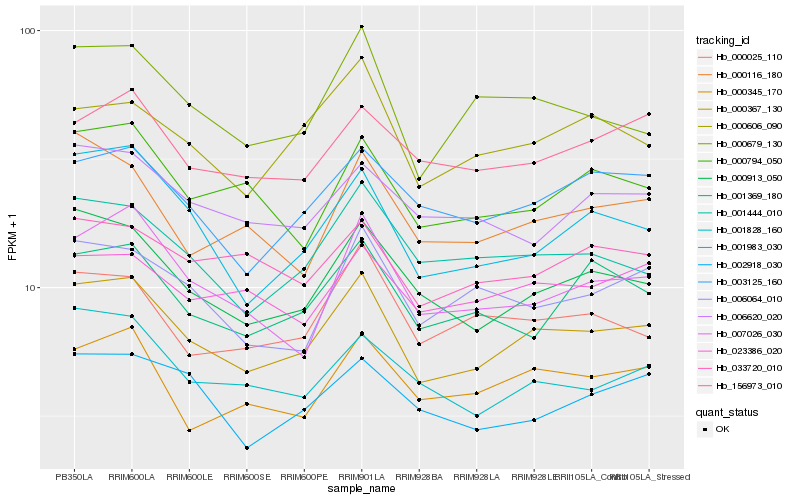
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000367_130 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_000913_050 |
0.062202258 |
- |
- |
hypothetical protein JCGZ_17329 [Jatropha curcas] |
| 3 |
Hb_000679_130 |
0.0721342355 |
- |
- |
dead box ATP-dependent RNA helicase, putative [Ricinus communis] |
| 4 |
Hb_001983_030 |
0.0735583954 |
transcription factor |
TF Family: FAR1 |
PREDICTED: protein FAR1-RELATED SEQUENCE 12 isoform X2 [Jatropha curcas] |
| 5 |
Hb_001444_010 |
0.0759612126 |
transcription factor |
TF Family: DDT |
PREDICTED: DDT domain-containing protein DDB_G0282237 isoform X1 [Jatropha curcas] |
| 6 |
Hb_007026_030 |
0.0775919274 |
- |
- |
rubisco subunit binding-protein alpha subunit, ruba, putative [Ricinus communis] |
| 7 |
Hb_001828_160 |
0.0778995143 |
- |
- |
hypothetical protein F775_31105 [Aegilops tauschii] |
| 8 |
Hb_003125_160 |
0.0788133137 |
transcription factor |
TF Family: NF-YC |
PREDICTED: nuclear transcription factor Y subunit C-1 isoform X1 [Jatropha curcas] |
| 9 |
Hb_001369_180 |
0.0798368531 |
- |
- |
PREDICTED: protein RFT1 homolog isoform X1 [Jatropha curcas] |
| 10 |
Hb_000606_090 |
0.079984198 |
- |
- |
Far upstream element-binding protein, putative [Ricinus communis] |
| 11 |
Hb_000794_050 |
0.081042878 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 12 [Jatropha curcas] |
| 12 |
Hb_006064_010 |
0.0811676596 |
- |
- |
PREDICTED: DDB1- and CUL4-associated factor 4 [Jatropha curcas] |
| 13 |
Hb_000025_110 |
0.0817289334 |
- |
- |
hypothetical protein JCGZ_22722 [Jatropha curcas] |
| 14 |
Hb_000345_170 |
0.0819687117 |
- |
- |
PREDICTED: lysine-specific demethylase JMJ25 isoform X2 [Jatropha curcas] |
| 15 |
Hb_033720_010 |
0.0844582621 |
- |
- |
PREDICTED: uncharacterized protein LOC105648843 [Jatropha curcas] |
| 16 |
Hb_156973_010 |
0.0850012474 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_000116_180 |
0.0851089499 |
- |
- |
PREDICTED: heterogeneous nuclear ribonucleoprotein H isoform X1 [Jatropha curcas] |
| 18 |
Hb_002918_030 |
0.0852722626 |
- |
- |
PREDICTED: dol-P-Glc:Glc(2)Man(9)GlcNAc(2)-PP-Dol alpha-1,2-glucosyltransferase isoform X1 [Jatropha curcas] |
| 19 |
Hb_023386_020 |
0.0865644921 |
- |
- |
PREDICTED: chaperone protein dnaJ 13 isoform X2 [Jatropha curcas] |
| 20 |
Hb_006620_020 |
0.0870776074 |
- |
- |
PREDICTED: uncharacterized protein LOC105638878 [Jatropha curcas] |