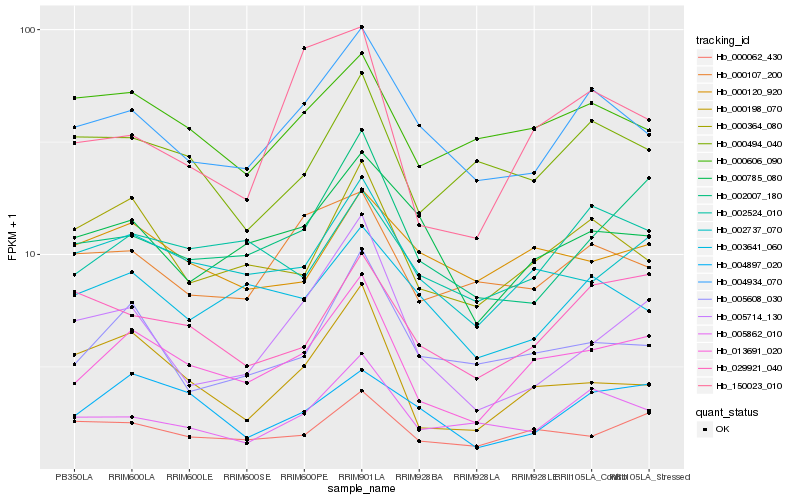
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_013691_020 |
0.0 |
- |
- |
transcription initiation factor iie, alpha subunit, putative [Ricinus communis] |
| 2 |
Hb_002737_070 |
0.0991800506 |
- |
- |
PREDICTED: phagocyte signaling-impaired protein [Jatropha curcas] |
| 3 |
Hb_000062_430 |
0.1221878392 |
- |
- |
PREDICTED: uncharacterized protein LOC105644764 isoform X1 [Jatropha curcas] |
| 4 |
Hb_000364_080 |
0.1254537566 |
- |
- |
PREDICTED: OTU domain-containing protein 3 isoform X1 [Jatropha curcas] |
| 5 |
Hb_002007_180 |
0.1256600347 |
- |
- |
PREDICTED: uncharacterized protein LOC105647525 [Jatropha curcas] |
| 6 |
Hb_000198_070 |
0.1257101533 |
- |
- |
PREDICTED: cyclin-dependent kinase inhibitor 7-like [Jatropha curcas] |
| 7 |
Hb_005608_030 |
0.1265978357 |
- |
- |
PREDICTED: uncharacterized protein LOC104110884 [Nicotiana tomentosiformis] |
| 8 |
Hb_004934_070 |
0.1344400651 |
- |
- |
PREDICTED: U2 small nuclear ribonucleoprotein A' [Jatropha curcas] |
| 9 |
Hb_000785_080 |
0.1386162065 |
- |
- |
PREDICTED: zinc finger CCCH domain-containing protein 13 [Jatropha curcas] |
| 10 |
Hb_002524_010 |
0.1409045233 |
- |
- |
hypothetical protein POPTR_0006s20250g [Populus trichocarpa] |
| 11 |
Hb_005862_010 |
0.1430408637 |
- |
- |
PREDICTED: riboflavin biosynthesis protein PYRD, chloroplastic [Jatropha curcas] |
| 12 |
Hb_004897_020 |
0.1445441596 |
- |
- |
PREDICTED: (-)-germacrene D synthase-like [Populus euphratica] |
| 13 |
Hb_000606_090 |
0.1459290775 |
- |
- |
Far upstream element-binding protein, putative [Ricinus communis] |
| 14 |
Hb_000120_920 |
0.1480089088 |
- |
- |
poly(A) polymerase, putative [Ricinus communis] |
| 15 |
Hb_029921_040 |
0.1496370039 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_003641_060 |
0.1501838124 |
- |
- |
PREDICTED: uncharacterized protein LOC105638019 [Jatropha curcas] |
| 17 |
Hb_000107_200 |
0.1513660804 |
- |
- |
hypothetical protein B456_005G214800 [Gossypium raimondii] |
| 18 |
Hb_000494_040 |
0.1514810212 |
- |
- |
PREDICTED: putative dual specificity protein phosphatase DSP8 [Jatropha curcas] |
| 19 |
Hb_005714_130 |
0.1516122861 |
transcription factor |
TF Family: C2H2 |
unknown [Populus trichocarpa x Populus deltoides] |
| 20 |
Hb_150023_010 |
0.1517928128 |
- |
- |
50S ribosomal protein L2, putative [Ricinus communis] |