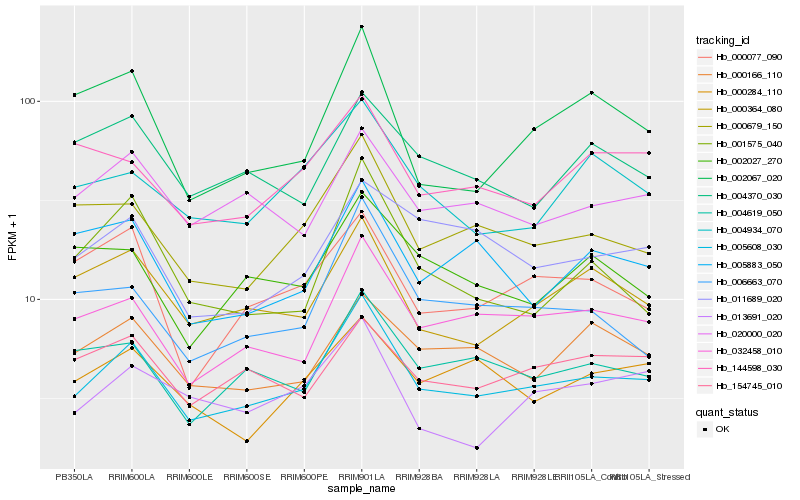
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005608_030 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC104110884 [Nicotiana tomentosiformis] |
| 2 |
Hb_032458_010 |
0.0869606572 |
- |
- |
hypothetical protein JCGZ_21107 [Jatropha curcas] |
| 3 |
Hb_000679_150 |
0.1022684206 |
- |
- |
PREDICTED: DEAD-box ATP-dependent RNA helicase 56 isoform X2 [Fragaria vesca subsp. vesca] |
| 4 |
Hb_004619_050 |
0.1063967617 |
- |
- |
PREDICTED: phosphatidylinositol 4-phosphate 5-kinase 8-like isoform X2 [Jatropha curcas] |
| 5 |
Hb_000364_080 |
0.1079820735 |
- |
- |
PREDICTED: OTU domain-containing protein 3 isoform X1 [Jatropha curcas] |
| 6 |
Hb_020000_020 |
0.1125777586 |
- |
- |
Troponin C, skeletal muscle, putative [Ricinus communis] |
| 7 |
Hb_006663_070 |
0.1145139491 |
- |
- |
PREDICTED: uncharacterized protein LOC105637432 [Jatropha curcas] |
| 8 |
Hb_004934_070 |
0.11649186 |
- |
- |
PREDICTED: U2 small nuclear ribonucleoprotein A' [Jatropha curcas] |
| 9 |
Hb_000166_110 |
0.1184800914 |
transcription factor |
TF Family: SNF2 |
PREDICTED: protein CHROMATIN REMODELING 25 [Jatropha curcas] |
| 10 |
Hb_000077_090 |
0.1198072205 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_002067_020 |
0.1212181336 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_001575_040 |
0.1212409794 |
- |
- |
PREDICTED: U3 small nucleolar RNA-associated protein 25 isoform X2 [Jatropha curcas] |
| 13 |
Hb_154745_010 |
0.1223043658 |
- |
- |
dead box ATP-dependent RNA helicase, putative [Ricinus communis] |
| 14 |
Hb_005883_050 |
0.1226297777 |
- |
- |
PREDICTED: transportin-3 [Jatropha curcas] |
| 15 |
Hb_011689_020 |
0.1248676872 |
- |
- |
protein phosphatase 2c, putative [Ricinus communis] |
| 16 |
Hb_000284_110 |
0.1250156729 |
- |
- |
PREDICTED: bromodomain-containing protein 3 [Jatropha curcas] |
| 17 |
Hb_002027_270 |
0.125142623 |
- |
- |
PREDICTED: serine/threonine-protein phosphatase BSL1 [Jatropha curcas] |
| 18 |
Hb_004370_030 |
0.1257126061 |
- |
- |
PREDICTED: RNA-binding protein 34 [Jatropha curcas] |
| 19 |
Hb_144598_030 |
0.1264010252 |
- |
- |
PREDICTED: mitotic apparatus protein p62 [Jatropha curcas] |
| 20 |
Hb_013691_020 |
0.1265978357 |
- |
- |
transcription initiation factor iie, alpha subunit, putative [Ricinus communis] |