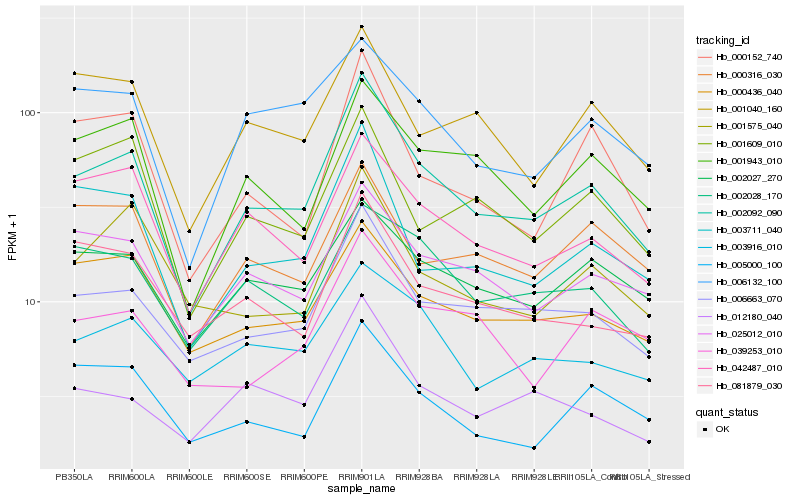
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002092_090 |
0.0 |
- |
- |
PREDICTED: aspartic proteinase Asp1 [Jatropha curcas] |
| 2 |
Hb_006663_070 |
0.0943987742 |
- |
- |
PREDICTED: uncharacterized protein LOC105637432 [Jatropha curcas] |
| 3 |
Hb_025012_010 |
0.1153237054 |
- |
- |
PREDICTED: importin-4 [Vitis vinifera] |
| 4 |
Hb_042487_010 |
0.1159209353 |
- |
- |
PREDICTED: DEAD-box ATP-dependent RNA helicase 9-like [Jatropha curcas] |
| 5 |
Hb_003711_040 |
0.1170808731 |
- |
- |
PREDICTED: UBX domain-containing protein 4 [Jatropha curcas] |
| 6 |
Hb_001943_010 |
0.1174416434 |
- |
- |
PREDICTED: heterogeneous nuclear ribonucleoprotein 1 [Jatropha curcas] |
| 7 |
Hb_012180_040 |
0.1210176448 |
- |
- |
hypothetical protein VITISV_042517 [Vitis vinifera] |
| 8 |
Hb_002027_270 |
0.1236920093 |
- |
- |
PREDICTED: serine/threonine-protein phosphatase BSL1 [Jatropha curcas] |
| 9 |
Hb_006132_100 |
0.126145189 |
- |
- |
PREDICTED: uncharacterized protein LOC105642968 [Jatropha curcas] |
| 10 |
Hb_003916_010 |
0.1263914761 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 13 [Jatropha curcas] |
| 11 |
Hb_000152_740 |
0.1266856197 |
- |
- |
Cisplatin resistance-associated overexpressed protein, putative [Ricinus communis] |
| 12 |
Hb_000436_040 |
0.1267863475 |
- |
- |
PREDICTED: transportin-1 [Jatropha curcas] |
| 13 |
Hb_001040_160 |
0.1283062977 |
- |
- |
glutaredoxin, grx, putative [Ricinus communis] |
| 14 |
Hb_001575_040 |
0.1295752352 |
- |
- |
PREDICTED: U3 small nucleolar RNA-associated protein 25 isoform X2 [Jatropha curcas] |
| 15 |
Hb_002028_170 |
0.1297681021 |
- |
- |
PREDICTED: brefeldin A-inhibited guanine nucleotide-exchange protein 5 [Jatropha curcas] |
| 16 |
Hb_081879_030 |
0.1310497434 |
- |
- |
PREDICTED: U3 small nucleolar RNA-associated protein 21 homolog [Jatropha curcas] |
| 17 |
Hb_039253_010 |
0.1313265384 |
- |
- |
PREDICTED: zinc finger CCCH domain-containing protein 31 isoform X2 [Jatropha curcas] |
| 18 |
Hb_001609_010 |
0.1330432744 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: putative inactive disease susceptibility protein LOV1 [Jatropha curcas] |
| 19 |
Hb_005000_100 |
0.1335868839 |
- |
- |
PCI domain-containing protein 2 [Morus notabilis] |
| 20 |
Hb_000316_030 |
0.1342645953 |
- |
- |
zinc finger protein, putative [Ricinus communis] |