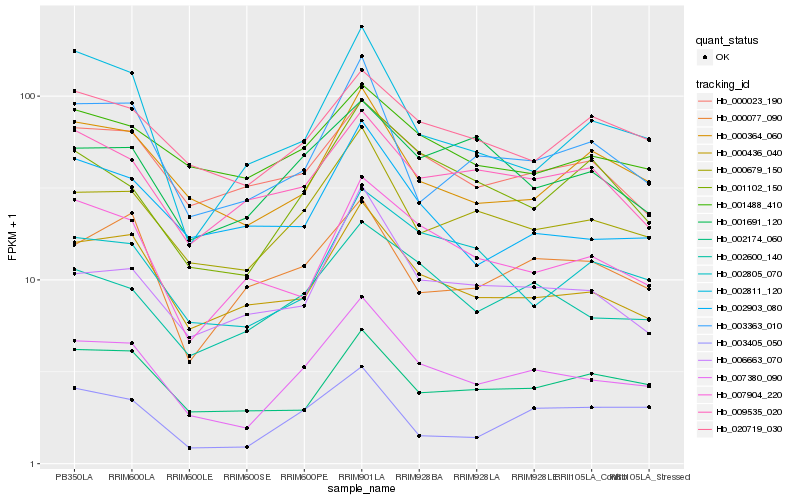
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_007380_090 |
0.0 |
- |
- |
- |
| 2 |
Hb_000679_150 |
0.0897203168 |
- |
- |
PREDICTED: DEAD-box ATP-dependent RNA helicase 56 isoform X2 [Fragaria vesca subsp. vesca] |
| 3 |
Hb_000436_040 |
0.1035191804 |
- |
- |
PREDICTED: transportin-1 [Jatropha curcas] |
| 4 |
Hb_002600_140 |
0.1041154437 |
- |
- |
PREDICTED: protein FLX-like 2 [Jatropha curcas] |
| 5 |
Hb_000364_060 |
0.1160840529 |
- |
- |
JHL03K20.4 [Jatropha curcas] |
| 6 |
Hb_003363_010 |
0.117249832 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_001102_150 |
0.1174335693 |
- |
- |
PREDICTED: ethanolamine-phosphate cytidylyltransferase-like isoform X1 [Jatropha curcas] |
| 8 |
Hb_006663_070 |
0.1204491737 |
- |
- |
PREDICTED: uncharacterized protein LOC105637432 [Jatropha curcas] |
| 9 |
Hb_000023_190 |
0.1255282793 |
- |
- |
PREDICTED: eukaryotic translation initiation factor 3 subunit B-like [Jatropha curcas] |
| 10 |
Hb_020719_030 |
0.1260046149 |
- |
- |
PREDICTED: ER membrane protein complex subunit 3-like [Jatropha curcas] |
| 11 |
Hb_002174_060 |
0.1269214781 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g09900-like [Jatropha curcas] |
| 12 |
Hb_002805_070 |
0.1279823676 |
transcription factor |
TF Family: HB |
PREDICTED: homeobox protein 10 isoform X2 [Jatropha curcas] |
| 13 |
Hb_002903_080 |
0.1303624093 |
- |
- |
PREDICTED: splicing factor 3B subunit 2 [Jatropha curcas] |
| 14 |
Hb_007904_220 |
0.1310158298 |
- |
- |
PREDICTED: protein SDA1 homolog isoform X1 [Jatropha curcas] |
| 15 |
Hb_002811_120 |
0.131327599 |
- |
- |
PREDICTED: DNA-damage-repair/toleration protein DRT102 [Jatropha curcas] |
| 16 |
Hb_001488_410 |
0.131395123 |
- |
- |
PREDICTED: eukaryotic translation initiation factor 3 subunit M [Jatropha curcas] |
| 17 |
Hb_001691_120 |
0.131737502 |
desease resistance |
Gene Name: AAA |
PREDICTED: vesicle-fusing ATPase [Jatropha curcas] |
| 18 |
Hb_003405_050 |
0.131914735 |
- |
- |
- |
| 19 |
Hb_009535_020 |
0.1319410465 |
- |
- |
PREDICTED: V-type proton ATPase subunit a3-like [Jatropha curcas] |
| 20 |
Hb_000077_090 |
0.1325996042 |
- |
- |
conserved hypothetical protein [Ricinus communis] |