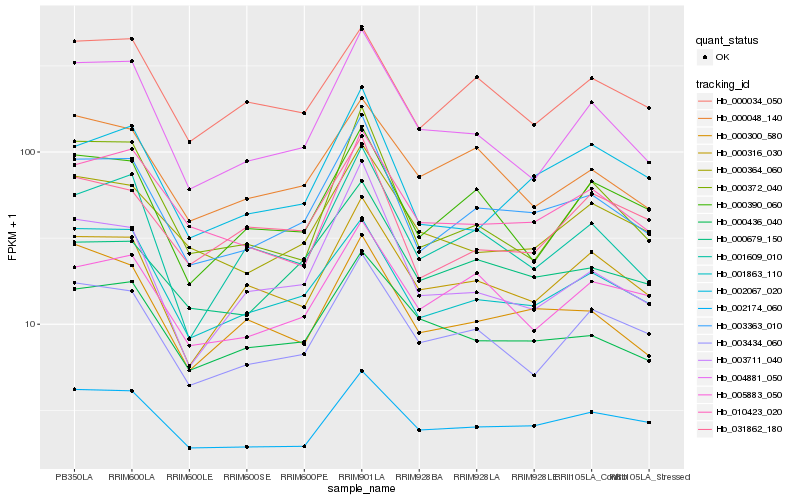
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003363_010 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_001863_110 |
0.0801383683 |
- |
- |
PREDICTED: uncharacterized protein At1g04910 isoform X1 [Jatropha curcas] |
| 3 |
Hb_000679_150 |
0.0899308135 |
- |
- |
PREDICTED: DEAD-box ATP-dependent RNA helicase 56 isoform X2 [Fragaria vesca subsp. vesca] |
| 4 |
Hb_003434_060 |
0.0903675534 |
- |
- |
PREDICTED: membrane-bound transcription factor site-2 protease homolog isoform X1 [Jatropha curcas] |
| 5 |
Hb_001609_010 |
0.0914358217 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: putative inactive disease susceptibility protein LOV1 [Jatropha curcas] |
| 6 |
Hb_000316_030 |
0.0916788355 |
- |
- |
zinc finger protein, putative [Ricinus communis] |
| 7 |
Hb_002174_060 |
0.0922439712 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g09900-like [Jatropha curcas] |
| 8 |
Hb_004881_050 |
0.094491983 |
- |
- |
PREDICTED: obg-like ATPase 1 [Jatropha curcas] |
| 9 |
Hb_000364_060 |
0.0951967347 |
- |
- |
JHL03K20.4 [Jatropha curcas] |
| 10 |
Hb_000300_580 |
0.0957278513 |
- |
- |
PREDICTED: importin-5-like [Jatropha curcas] |
| 11 |
Hb_002067_020 |
0.0965883055 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_000390_060 |
0.0983541842 |
- |
- |
PREDICTED: F-box/kelch-repeat protein At5g60570-like [Jatropha curcas] |
| 13 |
Hb_000436_040 |
0.0985180505 |
- |
- |
PREDICTED: transportin-1 [Jatropha curcas] |
| 14 |
Hb_005883_050 |
0.0994773925 |
- |
- |
PREDICTED: transportin-3 [Jatropha curcas] |
| 15 |
Hb_000034_050 |
0.0998970416 |
- |
- |
PREDICTED: uncharacterized protein LOC105637912 [Jatropha curcas] |
| 16 |
Hb_010423_020 |
0.1004557627 |
- |
- |
PREDICTED: pre-mRNA-splicing factor CWC25 homolog [Jatropha curcas] |
| 17 |
Hb_031862_180 |
0.1011237083 |
- |
- |
PREDICTED: uncharacterized protein LOC105644669 [Jatropha curcas] |
| 18 |
Hb_003711_040 |
0.1012550213 |
- |
- |
PREDICTED: UBX domain-containing protein 4 [Jatropha curcas] |
| 19 |
Hb_000048_140 |
0.1023489394 |
- |
- |
PREDICTED: eukaryotic peptide chain release factor GTP-binding subunit ERF3A isoform X3 [Jatropha curcas] |
| 20 |
Hb_000372_040 |
0.1034364958 |
- |
- |
WD-repeat protein, putative [Ricinus communis] |