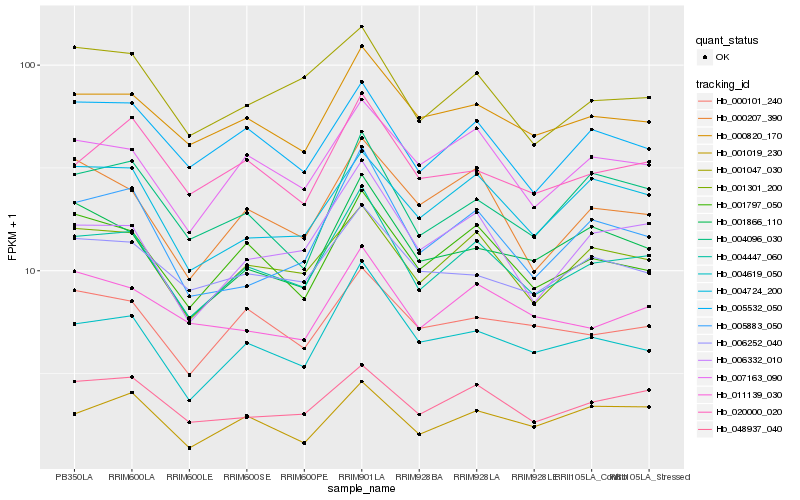
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004447_060 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105636329 isoform X2 [Jatropha curcas] |
| 2 |
Hb_001301_200 |
0.0569968612 |
desease resistance |
Gene Name: NB-ARC |
Disease resistance protein RPS2, putative [Ricinus communis] |
| 3 |
Hb_001797_050 |
0.0657214796 |
transcription factor |
TF Family: FAR1 |
PREDICTED: protein FAR1-RELATED SEQUENCE 3-like isoform X1 [Jatropha curcas] |
| 4 |
Hb_004619_050 |
0.0659821831 |
- |
- |
PREDICTED: phosphatidylinositol 4-phosphate 5-kinase 8-like isoform X2 [Jatropha curcas] |
| 5 |
Hb_048937_040 |
0.0669279981 |
- |
- |
APO protein 4, mitochondrial precursor, putative [Ricinus communis] |
| 6 |
Hb_007163_090 |
0.0670937642 |
- |
- |
PREDICTED: protein spt2-like [Jatropha curcas] |
| 7 |
Hb_000820_170 |
0.0676986594 |
- |
- |
PREDICTED: THO complex subunit 4A [Jatropha curcas] |
| 8 |
Hb_006332_010 |
0.0698825747 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At5g10290 isoform X1 [Vitis vinifera] |
| 9 |
Hb_005532_050 |
0.0741912065 |
- |
- |
PREDICTED: probable leucine-rich repeat receptor-like protein kinase At1g35710 [Jatropha curcas] |
| 10 |
Hb_005883_050 |
0.0759760619 |
- |
- |
PREDICTED: transportin-3 [Jatropha curcas] |
| 11 |
Hb_001047_030 |
0.079396987 |
- |
- |
PREDICTED: protein transport protein SEC13 homolog B [Jatropha curcas] |
| 12 |
Hb_004096_030 |
0.0801140571 |
- |
- |
PREDICTED: KH domain-containing protein At4g18375-like isoform X1 [Jatropha curcas] |
| 13 |
Hb_000101_240 |
0.0803043856 |
desease resistance |
Gene Name: NB-ARC |
putative disease resistance gene NBS-LRR family protein [Populus trichocarpa] |
| 14 |
Hb_020000_020 |
0.0808063767 |
- |
- |
Troponin C, skeletal muscle, putative [Ricinus communis] |
| 15 |
Hb_001866_110 |
0.0813953535 |
- |
- |
PREDICTED: trihelix transcription factor GT-3b-like [Jatropha curcas] |
| 16 |
Hb_006252_040 |
0.0815916874 |
- |
- |
queuine tRNA-ribosyltransferase, putative [Ricinus communis] |
| 17 |
Hb_001019_230 |
0.0816705939 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g08070 [Jatropha curcas] |
| 18 |
Hb_000207_390 |
0.0819777581 |
- |
- |
PREDICTED: uncharacterized protein LOC105637143 [Jatropha curcas] |
| 19 |
Hb_004724_200 |
0.0828586788 |
- |
- |
PREDICTED: WD-40 repeat-containing protein MSI1 [Jatropha curcas] |
| 20 |
Hb_011139_030 |
0.0828661541 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase MBR2-like [Jatropha curcas] |