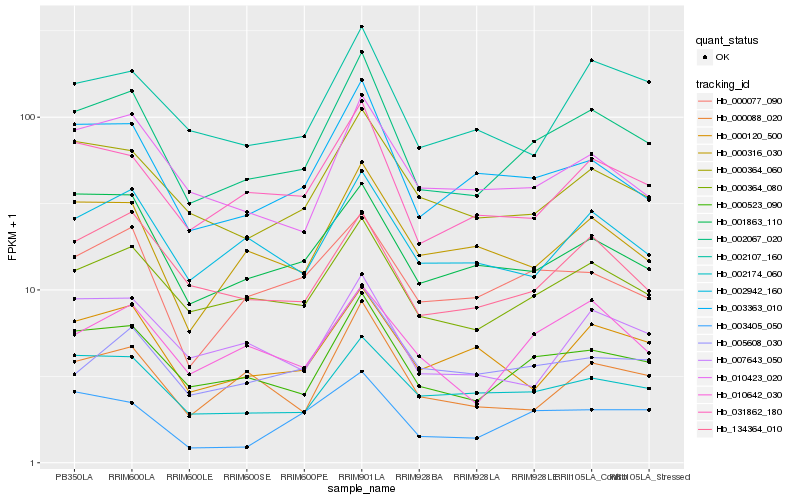
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002067_020 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_000523_090 |
0.0647536843 |
- |
- |
PREDICTED: protein DJ-1 homolog D-like [Populus euphratica] |
| 3 |
Hb_000364_080 |
0.0794126375 |
- |
- |
PREDICTED: OTU domain-containing protein 3 isoform X1 [Jatropha curcas] |
| 4 |
Hb_031862_180 |
0.0944285504 |
- |
- |
PREDICTED: uncharacterized protein LOC105644669 [Jatropha curcas] |
| 5 |
Hb_003363_010 |
0.0965883055 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_000364_060 |
0.103699477 |
- |
- |
JHL03K20.4 [Jatropha curcas] |
| 7 |
Hb_134364_010 |
0.1068217653 |
- |
- |
PREDICTED: transcriptional adapter ADA2b isoform X1 [Jatropha curcas] |
| 8 |
Hb_002174_060 |
0.1079034274 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g09900-like [Jatropha curcas] |
| 9 |
Hb_002942_160 |
0.1112123011 |
- |
- |
PREDICTED: coiled-coil domain-containing protein SCD2 [Jatropha curcas] |
| 10 |
Hb_010423_020 |
0.1113372218 |
- |
- |
PREDICTED: pre-mRNA-splicing factor CWC25 homolog [Jatropha curcas] |
| 11 |
Hb_010642_030 |
0.1130226629 |
- |
- |
zinc phosphodiesterase, putative [Ricinus communis] |
| 12 |
Hb_000077_090 |
0.1132701407 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_000316_030 |
0.1136327837 |
- |
- |
zinc finger protein, putative [Ricinus communis] |
| 14 |
Hb_002107_160 |
0.1137114269 |
- |
- |
PREDICTED: ribonuclease P protein subunit p25-like protein [Jatropha curcas] |
| 15 |
Hb_000088_020 |
0.1146289201 |
transcription factor |
TF Family: M-type |
hypothetical protein JCGZ_10946 [Jatropha curcas] |
| 16 |
Hb_001863_110 |
0.1152135188 |
- |
- |
PREDICTED: uncharacterized protein At1g04910 isoform X1 [Jatropha curcas] |
| 17 |
Hb_003405_050 |
0.1204124098 |
- |
- |
- |
| 18 |
Hb_000120_500 |
0.1211423779 |
- |
- |
PREDICTED: AT-hook motif nuclear-localized protein 10-like [Jatropha curcas] |
| 19 |
Hb_005608_030 |
0.1212181336 |
- |
- |
PREDICTED: uncharacterized protein LOC104110884 [Nicotiana tomentosiformis] |
| 20 |
Hb_007643_050 |
0.1218083515 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |