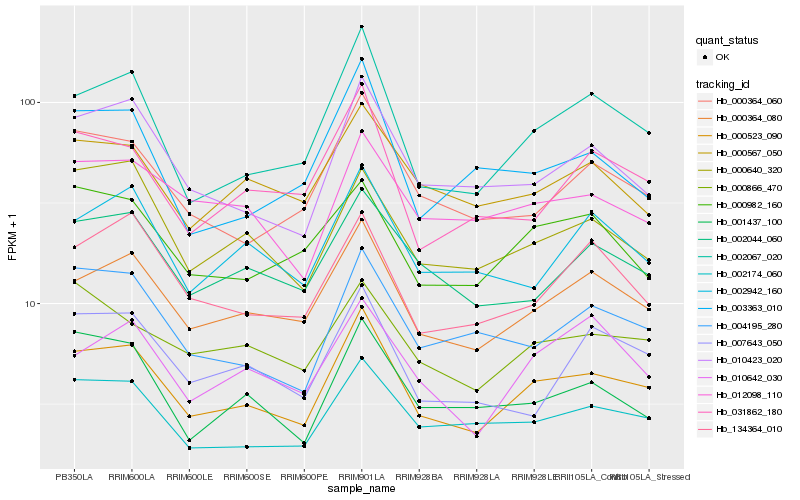
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000523_090 |
0.0 |
- |
- |
PREDICTED: protein DJ-1 homolog D-like [Populus euphratica] |
| 2 |
Hb_002067_020 |
0.0647536843 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_000364_080 |
0.0715115387 |
- |
- |
PREDICTED: OTU domain-containing protein 3 isoform X1 [Jatropha curcas] |
| 4 |
Hb_002174_060 |
0.0896976751 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g09900-like [Jatropha curcas] |
| 5 |
Hb_010423_020 |
0.089876672 |
- |
- |
PREDICTED: pre-mRNA-splicing factor CWC25 homolog [Jatropha curcas] |
| 6 |
Hb_000364_060 |
0.093156991 |
- |
- |
JHL03K20.4 [Jatropha curcas] |
| 7 |
Hb_004195_280 |
0.1002456601 |
- |
- |
PREDICTED: CDK5RAP1-like protein [Jatropha curcas] |
| 8 |
Hb_031862_180 |
0.1002766811 |
- |
- |
PREDICTED: uncharacterized protein LOC105644669 [Jatropha curcas] |
| 9 |
Hb_002044_060 |
0.1016391899 |
- |
- |
elongation factor ts, putative [Ricinus communis] |
| 10 |
Hb_134364_010 |
0.1052552991 |
- |
- |
PREDICTED: transcriptional adapter ADA2b isoform X1 [Jatropha curcas] |
| 11 |
Hb_000640_320 |
0.1059886937 |
- |
- |
PREDICTED: uncharacterized protein LOC105642644 isoform X2 [Jatropha curcas] |
| 12 |
Hb_000866_470 |
0.1065712957 |
- |
- |
PREDICTED: uncharacterized protein LOC105643015 [Jatropha curcas] |
| 13 |
Hb_002942_160 |
0.1072004878 |
- |
- |
PREDICTED: coiled-coil domain-containing protein SCD2 [Jatropha curcas] |
| 14 |
Hb_001437_100 |
0.1095127969 |
- |
- |
PREDICTED: DNA-binding protein REB1-like [Jatropha curcas] |
| 15 |
Hb_003363_010 |
0.1097442254 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_012098_110 |
0.1098142681 |
- |
- |
Ribonuclease 3 [Gossypium arboreum] |
| 17 |
Hb_000567_050 |
0.1100070195 |
- |
- |
PREDICTED: UBP1-associated protein 2C [Jatropha curcas] |
| 18 |
Hb_000982_160 |
0.1103195284 |
- |
- |
PREDICTED: CLIP-associated protein [Jatropha curcas] |
| 19 |
Hb_007643_050 |
0.1112797242 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 20 |
Hb_010642_030 |
0.1115029439 |
- |
- |
zinc phosphodiesterase, putative [Ricinus communis] |