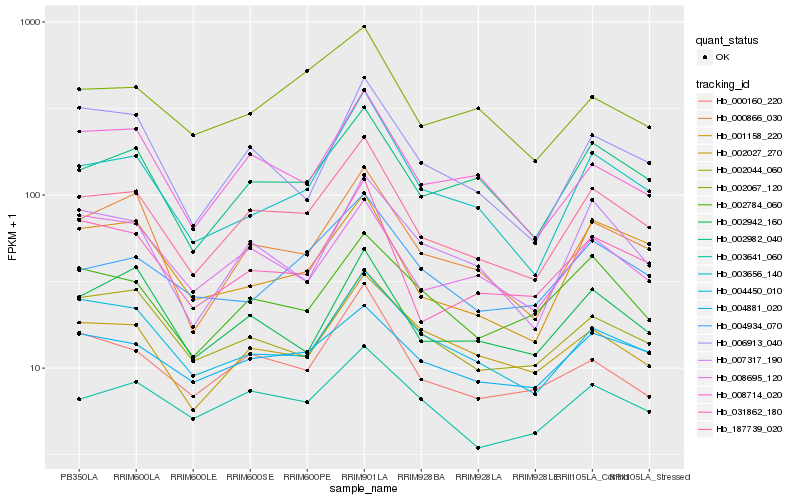
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_187739_020 |
0.0 |
- |
- |
PREDICTED: B2 protein [Jatropha curcas] |
| 2 |
Hb_000866_030 |
0.0748754212 |
- |
- |
hypothetical protein RCOM_1615820 [Ricinus communis] |
| 3 |
Hb_002982_040 |
0.0769618921 |
- |
- |
mta/sah nucleosidase, putative [Ricinus communis] |
| 4 |
Hb_003656_140 |
0.0818016876 |
- |
- |
PREDICTED: PHD finger protein ALFIN-LIKE 4 isoform X1 [Jatropha curcas] |
| 5 |
Hb_000160_220 |
0.0856651014 |
- |
- |
PREDICTED: protein AAR2 homolog isoform X1 [Jatropha curcas] |
| 6 |
Hb_031862_180 |
0.0858431433 |
- |
- |
PREDICTED: uncharacterized protein LOC105644669 [Jatropha curcas] |
| 7 |
Hb_008714_020 |
0.0889994136 |
- |
- |
PREDICTED: GTP-binding protein SAR1A-like [Jatropha curcas] |
| 8 |
Hb_003641_060 |
0.0910284482 |
- |
- |
PREDICTED: uncharacterized protein LOC105638019 [Jatropha curcas] |
| 9 |
Hb_004450_010 |
0.0927841006 |
- |
- |
PREDICTED: methyltransferase-like protein 10 [Jatropha curcas] |
| 10 |
Hb_002942_160 |
0.0947857843 |
- |
- |
PREDICTED: coiled-coil domain-containing protein SCD2 [Jatropha curcas] |
| 11 |
Hb_002027_270 |
0.0958457378 |
- |
- |
PREDICTED: serine/threonine-protein phosphatase BSL1 [Jatropha curcas] |
| 12 |
Hb_004881_020 |
0.096341016 |
- |
- |
PREDICTED: serine/threonine-protein kinase rio2-like [Jatropha curcas] |
| 13 |
Hb_002784_060 |
0.0977514747 |
transcription factor |
TF Family: bZIP |
PREDICTED: transcription factor TGA6 isoform X1 [Jatropha curcas] |
| 14 |
Hb_004934_070 |
0.0980370449 |
- |
- |
PREDICTED: U2 small nuclear ribonucleoprotein A' [Jatropha curcas] |
| 15 |
Hb_007317_190 |
0.0981271204 |
- |
- |
PREDICTED: WD-40 repeat-containing protein MSI1 [Jatropha curcas] |
| 16 |
Hb_001158_220 |
0.099269454 |
- |
- |
PREDICTED: probable protein S-acyltransferase 7 [Jatropha curcas] |
| 17 |
Hb_002067_120 |
0.0994852427 |
- |
- |
PREDICTED: uncharacterized protein LOC105111554 isoform X1 [Populus euphratica] |
| 18 |
Hb_006913_040 |
0.1000846934 |
- |
- |
PREDICTED: hydroxyacylglutathione hydrolase cytoplasmic [Jatropha curcas] |
| 19 |
Hb_002044_060 |
0.1004855913 |
- |
- |
elongation factor ts, putative [Ricinus communis] |
| 20 |
Hb_008695_120 |
0.1008351716 |
transcription factor |
TF Family: C2H2 |
PREDICTED: MYST-like histone acetyltransferase 2 [Jatropha curcas] |