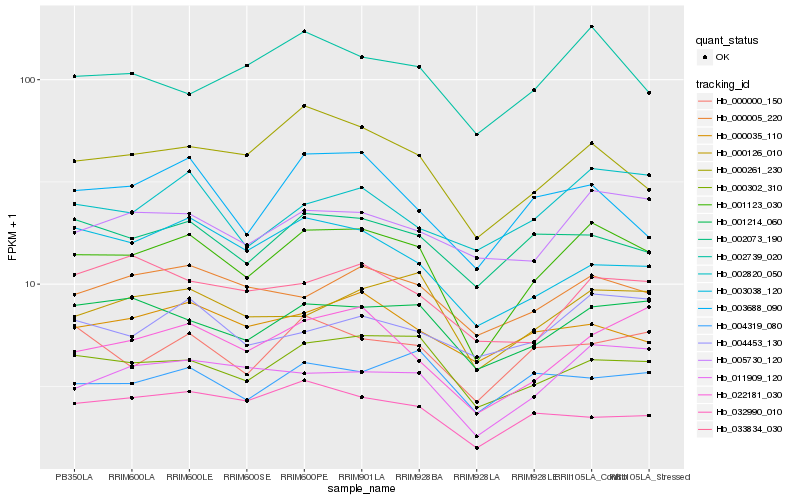
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001123_030 |
0.0 |
- |
- |
ABC transporter family protein [Hevea brasiliensis] |
| 2 |
Hb_000302_310 |
0.0858244398 |
- |
- |
PREDICTED: glutaminyl-peptide cyclotransferase-like [Jatropha curcas] |
| 3 |
Hb_000261_230 |
0.0911945356 |
- |
- |
PREDICTED: UBX domain-containing protein 4 [Vitis vinifera] |
| 4 |
Hb_001214_060 |
0.0920510045 |
- |
- |
PREDICTED: phosphoinositide phosphatase SAC8 [Jatropha curcas] |
| 5 |
Hb_011909_120 |
0.0937635939 |
- |
- |
Ubiquitin carboxyl-terminal hydrolase, putative [Ricinus communis] |
| 6 |
Hb_000126_010 |
0.0938721415 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase RFK1 isoform X1 [Jatropha curcas] |
| 7 |
Hb_022181_030 |
0.0986122789 |
- |
- |
PREDICTED: uncharacterized protein LOC105649615 isoform X2 [Jatropha curcas] |
| 8 |
Hb_000005_220 |
0.0987405306 |
- |
- |
DNA ligase 1, partial [Glycine soja] |
| 9 |
Hb_002073_190 |
0.0993530743 |
- |
- |
PREDICTED: uncharacterized protein LOC105649812 isoform X1 [Jatropha curcas] |
| 10 |
Hb_000000_150 |
0.0997425394 |
- |
- |
PREDICTED: uncharacterized protein LOC105633730 [Jatropha curcas] |
| 11 |
Hb_005730_120 |
0.1009821992 |
- |
- |
PREDICTED: protein WHI4 [Jatropha curcas] |
| 12 |
Hb_002739_020 |
0.1052763821 |
- |
- |
PREDICTED: UTP--glucose-1-phosphate uridylyltransferase [Jatropha curcas] |
| 13 |
Hb_000035_110 |
0.1071830737 |
- |
- |
PREDICTED: transcription initiation factor TFIID subunit 5 [Jatropha curcas] |
| 14 |
Hb_003688_090 |
0.1075502015 |
- |
- |
PREDICTED: ATP synthase subunit gamma, mitochondrial [Jatropha curcas] |
| 15 |
Hb_002820_050 |
0.1080382635 |
- |
- |
PREDICTED: putative glutathione-specific gamma-glutamylcyclotransferase 2 [Jatropha curcas] |
| 16 |
Hb_032990_010 |
0.1098363238 |
- |
- |
PREDICTED: fanconi-associated nuclease 1 homolog [Jatropha curcas] |
| 17 |
Hb_033834_030 |
0.1103327853 |
- |
- |
PREDICTED: nucleolar complex protein 2 homolog isoform X1 [Jatropha curcas] |
| 18 |
Hb_004453_130 |
0.1108937839 |
- |
- |
PREDICTED: tRNA (cytosine-5-)-methyltransferase isoform X1 [Jatropha curcas] |
| 19 |
Hb_003038_120 |
0.1109576226 |
- |
- |
rer1 protein, putative [Ricinus communis] |
| 20 |
Hb_004319_080 |
0.1113702628 |
- |
- |
PREDICTED: GPI ethanolamine phosphate transferase 1 [Jatropha curcas] |