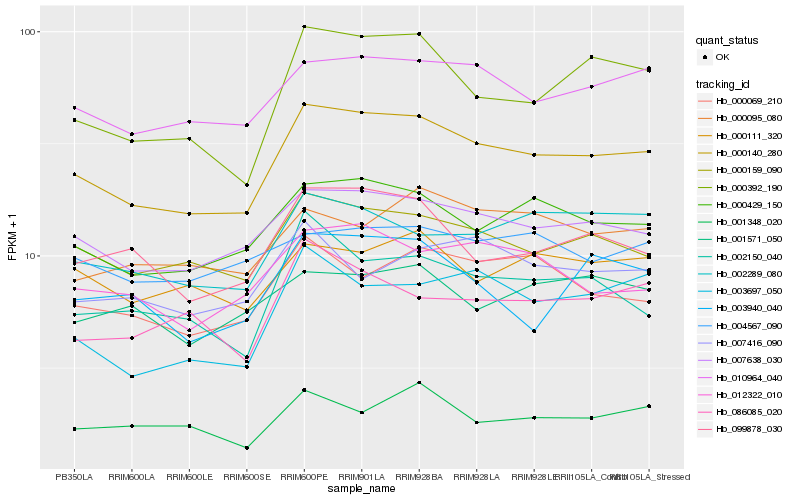
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000140_280 |
0.0 |
- |
- |
hypothetical protein B456_001G157900 [Gossypium raimondii] |
| 2 |
Hb_007638_030 |
0.0557517222 |
- |
- |
PREDICTED: phosphatidylinositol 3,4,5-trisphosphate 3-phosphatase and dual-specificity protein phosphatase PTEN [Jatropha curcas] |
| 3 |
Hb_000429_150 |
0.0621946187 |
- |
- |
hypothetical protein JCGZ_06799 [Jatropha curcas] |
| 4 |
Hb_000159_090 |
0.0636508295 |
- |
- |
hypothetical protein POPTR_0015s153201g, partial [Populus trichocarpa] |
| 5 |
Hb_010964_040 |
0.0696280375 |
- |
- |
Eukaryotic initiation factor 4A-3 [Gossypium arboreum] |
| 6 |
Hb_000392_190 |
0.0743412345 |
- |
- |
PREDICTED: NAP1-related protein 2 [Jatropha curcas] |
| 7 |
Hb_099878_030 |
0.0779808802 |
- |
- |
PREDICTED: coiled-coil domain-containing protein R3HCC1L isoform X4 [Jatropha curcas] |
| 8 |
Hb_012322_010 |
0.0807253917 |
- |
- |
Aspartic proteinase nepenthesin-1 precursor, putative [Ricinus communis] |
| 9 |
Hb_000069_210 |
0.0841411562 |
- |
- |
PREDICTED: exportin-2 [Jatropha curcas] |
| 10 |
Hb_002289_080 |
0.0856857241 |
- |
- |
hypothetical protein POPTR_0016s05590g [Populus trichocarpa] |
| 11 |
Hb_000111_320 |
0.0895097094 |
transcription factor |
TF Family: bZIP |
Transcription factor HBP-1b(c1), putative [Ricinus communis] |
| 12 |
Hb_002150_040 |
0.0899490558 |
- |
- |
PREDICTED: CMP-sialic acid transporter 1 isoform X3 [Jatropha curcas] |
| 13 |
Hb_003940_040 |
0.0928719123 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_000095_080 |
0.0931644823 |
- |
- |
PREDICTED: probable U3 small nucleolar RNA-associated protein 7 [Jatropha curcas] |
| 15 |
Hb_086085_020 |
0.0934625801 |
- |
- |
Non-imprinted in Prader-Willi/Angelman syndrome region protein isoform 3 [Theobroma cacao] |
| 16 |
Hb_003697_050 |
0.0941387682 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 13 [Jatropha curcas] |
| 17 |
Hb_001348_020 |
0.0950110253 |
- |
- |
PREDICTED: uncharacterized protein LOC105638926 isoform X2 [Jatropha curcas] |
| 18 |
Hb_007416_090 |
0.0963794697 |
- |
- |
UDP-sugar transporter, putative [Ricinus communis] |
| 19 |
Hb_004567_090 |
0.0964280447 |
- |
- |
PREDICTED: GPI transamidase component PIG-T [Jatropha curcas] |
| 20 |
Hb_001571_050 |
0.0969082602 |
- |
- |
PREDICTED: endoplasmic reticulum-Golgi intermediate compartment protein 3-like [Populus euphratica] |