| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
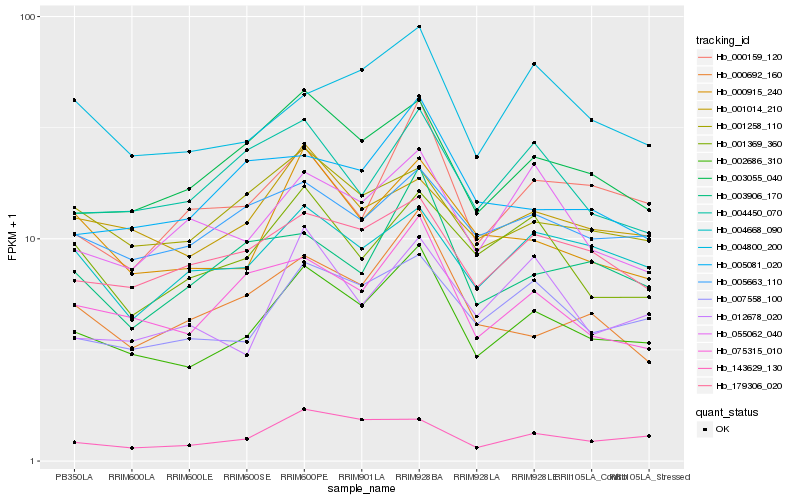
Hb_002686_310 |
0.0 |
- |
- |
PREDICTED: aquaporin SIP1-1 [Jatropha curcas] |
| 2 |
Hb_007558_100 |
0.0873108824 |
- |
- |
PREDICTED: uncharacterized protein LOC105649092 isoform X2 [Jatropha curcas] |
| 3 |
Hb_075315_010 |
0.0938652529 |
- |
- |
ornithine aminotransferase [Camellia sinensis] |
| 4 |
Hb_003055_040 |
0.0978737195 |
- |
- |
PREDICTED: secretory carrier-associated membrane protein 3-like isoform X1 [Jatropha curcas] |
| 5 |
Hb_000915_240 |
0.0987805189 |
- |
- |
catalytic, putative [Ricinus communis] |
| 6 |
Hb_179306_020 |
0.1018297236 |
- |
- |
PREDICTED: uncharacterized protein LOC105634392 isoform X1 [Jatropha curcas] |
| 7 |
Hb_143629_130 |
0.1033594069 |
- |
- |
unnamed protein product [Coffea canephora] |
| 8 |
Hb_000159_120 |
0.1086093813 |
- |
- |
PREDICTED: uncharacterized protein LOC105632857 [Jatropha curcas] |
| 9 |
Hb_005663_110 |
0.1087329874 |
- |
- |
PREDICTED: large proline-rich protein BAG6 isoform X2 [Jatropha curcas] |
| 10 |
Hb_001258_110 |
0.1090170416 |
- |
- |
transporter, putative [Ricinus communis] |
| 11 |
Hb_001014_210 |
0.1094332996 |
- |
- |
CYP51 [Hevea brasiliensis] |
| 12 |
Hb_004800_200 |
0.1113799148 |
- |
- |
- |
| 13 |
Hb_004668_090 |
0.1113962443 |
- |
- |
PREDICTED: uncharacterized protein LOC105630440 [Jatropha curcas] |
| 14 |
Hb_001369_360 |
0.112213504 |
- |
- |
embryo yellow protein, partial [Manihot esculenta] |
| 15 |
Hb_000692_160 |
0.1135411271 |
- |
- |
Transportin 1 isoform 1 [Theobroma cacao] |
| 16 |
Hb_004450_070 |
0.1155340503 |
- |
- |
PREDICTED: 2-hydroxyacyl-CoA lyase [Jatropha curcas] |
| 17 |
Hb_003906_170 |
0.1173829917 |
- |
- |
alcohol dehydrogenase, putative [Ricinus communis] |
| 18 |
Hb_055062_040 |
0.1182163459 |
- |
- |
ceramidase, putative [Ricinus communis] |
| 19 |
Hb_012678_020 |
0.1185698588 |
- |
- |
PREDICTED: outer envelope protein 61 isoform X1 [Jatropha curcas] |
| 20 |
Hb_005081_020 |
0.1189352032 |
- |
- |
PREDICTED: ninja-family protein mc410 [Jatropha curcas] |