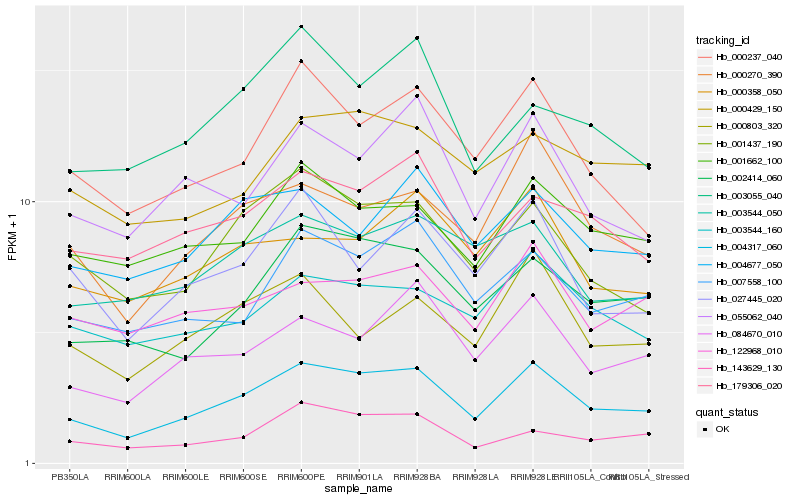
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004317_060 |
0.0 |
- |
- |
PREDICTED: testis-expressed sequence 10 protein isoform X2 [Jatropha curcas] |
| 2 |
Hb_002414_060 |
0.0879056184 |
- |
- |
hypothetical protein EUGRSUZ_E01310 [Eucalyptus grandis] |
| 3 |
Hb_000270_390 |
0.0950137155 |
- |
- |
PREDICTED: uncharacterized protein DDB_G0271670 [Jatropha curcas] |
| 4 |
Hb_001437_190 |
0.0970454494 |
- |
- |
ABC transporter family protein [Hevea brasiliensis] |
| 5 |
Hb_084670_010 |
0.0973201979 |
- |
- |
PREDICTED: thiamine biosynthetic bifunctional enzyme TH1, chloroplastic isoform X1 [Jatropha curcas] |
| 6 |
Hb_007558_100 |
0.0980895833 |
- |
- |
PREDICTED: uncharacterized protein LOC105649092 isoform X2 [Jatropha curcas] |
| 7 |
Hb_000237_040 |
0.0982463966 |
- |
- |
PREDICTED: V-type proton ATPase subunit a3-like [Jatropha curcas] |
| 8 |
Hb_001662_100 |
0.1034611119 |
- |
- |
PREDICTED: serine/threonine-protein kinase EDR1-like [Jatropha curcas] |
| 9 |
Hb_055062_040 |
0.1046892025 |
- |
- |
ceramidase, putative [Ricinus communis] |
| 10 |
Hb_003544_160 |
0.1073420935 |
- |
- |
- |
| 11 |
Hb_143629_130 |
0.1077507988 |
- |
- |
unnamed protein product [Coffea canephora] |
| 12 |
Hb_179306_020 |
0.1078712343 |
- |
- |
PREDICTED: uncharacterized protein LOC105634392 isoform X1 [Jatropha curcas] |
| 13 |
Hb_027445_020 |
0.1105302311 |
- |
- |
PREDICTED: glycosylphosphatidylinositol anchor attachment 1 protein [Jatropha curcas] |
| 14 |
Hb_004677_050 |
0.1105559285 |
- |
- |
PREDICTED: importin beta-like SAD2 isoform X2 [Jatropha curcas] |
| 15 |
Hb_000429_150 |
0.1112304123 |
- |
- |
hypothetical protein JCGZ_06799 [Jatropha curcas] |
| 16 |
Hb_003544_050 |
0.1115084725 |
- |
- |
PREDICTED: uncharacterized protein LOC105644300 isoform X1 [Jatropha curcas] |
| 17 |
Hb_000803_320 |
0.1116767662 |
transcription factor |
TF Family: PHD |
PREDICTED: histone-lysine N-methyltransferase ATX4 [Jatropha curcas] |
| 18 |
Hb_003055_040 |
0.1138974621 |
- |
- |
PREDICTED: secretory carrier-associated membrane protein 3-like isoform X1 [Jatropha curcas] |
| 19 |
Hb_000358_050 |
0.1139416615 |
- |
- |
PREDICTED: uncharacterized protein LOC105633378 [Jatropha curcas] |
| 20 |
Hb_122968_010 |
0.114578345 |
- |
- |
hypothetical protein JCGZ_23576 [Jatropha curcas] |