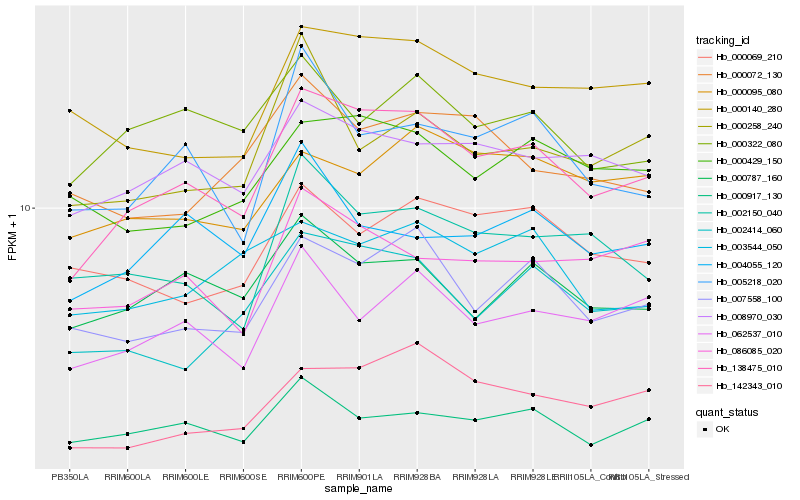
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_138475_010 |
0.0 |
- |
- |
PREDICTED: putative pentatricopeptide repeat-containing protein At1g12700, mitochondrial [Populus euphratica] |
| 2 |
Hb_007558_100 |
0.0938908293 |
- |
- |
PREDICTED: uncharacterized protein LOC105649092 isoform X2 [Jatropha curcas] |
| 3 |
Hb_008970_030 |
0.0997715889 |
- |
- |
arf gtpase-activating protein, putative [Ricinus communis] |
| 4 |
Hb_002414_060 |
0.1001159027 |
- |
- |
hypothetical protein EUGRSUZ_E01310 [Eucalyptus grandis] |
| 5 |
Hb_000917_130 |
0.101329061 |
- |
- |
PREDICTED: uncharacterized protein LOC105635730 [Jatropha curcas] |
| 6 |
Hb_062537_010 |
0.1049546804 |
- |
- |
hypothetical protein JCGZ_13884 [Jatropha curcas] |
| 7 |
Hb_000787_160 |
0.1079457256 |
- |
- |
PREDICTED: GDP-mannose transporter GONST1 isoform X1 [Jatropha curcas] |
| 8 |
Hb_086085_020 |
0.1093121844 |
- |
- |
Non-imprinted in Prader-Willi/Angelman syndrome region protein isoform 3 [Theobroma cacao] |
| 9 |
Hb_005218_020 |
0.110013533 |
- |
- |
Uncharacterized protein isoform 3 [Theobroma cacao] |
| 10 |
Hb_000140_280 |
0.1115872961 |
- |
- |
hypothetical protein B456_001G157900 [Gossypium raimondii] |
| 11 |
Hb_002150_040 |
0.1116737824 |
- |
- |
PREDICTED: CMP-sialic acid transporter 1 isoform X3 [Jatropha curcas] |
| 12 |
Hb_000429_150 |
0.1120527937 |
- |
- |
hypothetical protein JCGZ_06799 [Jatropha curcas] |
| 13 |
Hb_000095_080 |
0.1160333131 |
- |
- |
PREDICTED: probable U3 small nucleolar RNA-associated protein 7 [Jatropha curcas] |
| 14 |
Hb_003544_050 |
0.1169256261 |
- |
- |
PREDICTED: uncharacterized protein LOC105644300 isoform X1 [Jatropha curcas] |
| 15 |
Hb_000322_080 |
0.1173238389 |
- |
- |
hypothetical protein JCGZ_07423 [Jatropha curcas] |
| 16 |
Hb_000069_210 |
0.1197747432 |
- |
- |
PREDICTED: exportin-2 [Jatropha curcas] |
| 17 |
Hb_004055_120 |
0.1198743052 |
- |
- |
PREDICTED: uncharacterized protein LOC105631443 [Jatropha curcas] |
| 18 |
Hb_000072_130 |
0.1223739917 |
- |
- |
hypothetical protein OsJ_28567 [Oryza sativa Japonica Group] |
| 19 |
Hb_000258_240 |
0.1224294349 |
- |
- |
sur2 hydroxylase/desaturase, putative [Ricinus communis] |
| 20 |
Hb_142343_010 |
0.1225106992 |
- |
- |
ATP binding protein, putative [Ricinus communis] |