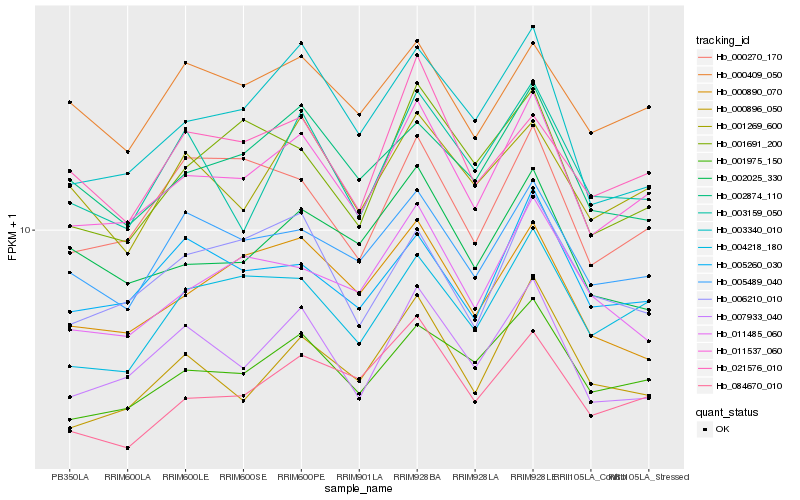
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_011537_060 |
0.0 |
- |
- |
UPF0061 protein azo1574 [Morus notabilis] |
| 2 |
Hb_001975_150 |
0.0619869139 |
- |
- |
PREDICTED: RINT1-like protein MAG2L [Jatropha curcas] |
| 3 |
Hb_007933_040 |
0.0688919602 |
- |
- |
PREDICTED: probable beta-D-xylosidase 6 isoform X2 [Jatropha curcas] |
| 4 |
Hb_004218_180 |
0.0729148525 |
- |
- |
PREDICTED: flowering time control protein FY isoform X2 [Jatropha curcas] |
| 5 |
Hb_005489_040 |
0.074810643 |
- |
- |
PREDICTED: cullin-1 [Jatropha curcas] |
| 6 |
Hb_003340_010 |
0.0773424091 |
- |
- |
PREDICTED: ubiquitin-activating enzyme E1 1 [Vitis vinifera] |
| 7 |
Hb_002874_110 |
0.0810019604 |
- |
- |
PREDICTED: uncharacterized protein LOC105637451 [Jatropha curcas] |
| 8 |
Hb_005260_030 |
0.0825948012 |
- |
- |
PREDICTED: uncharacterized protein LOC105633782 [Jatropha curcas] |
| 9 |
Hb_000270_170 |
0.0832342673 |
- |
- |
PREDICTED: formin-like protein 5 [Jatropha curcas] |
| 10 |
Hb_084670_010 |
0.083409405 |
- |
- |
PREDICTED: thiamine biosynthetic bifunctional enzyme TH1, chloroplastic isoform X1 [Jatropha curcas] |
| 11 |
Hb_002025_330 |
0.0844956869 |
- |
- |
peptide transporter, putative [Ricinus communis] |
| 12 |
Hb_000890_070 |
0.0850504333 |
- |
- |
PREDICTED: protein ALWAYS EARLY 3 isoform X2 [Jatropha curcas] |
| 13 |
Hb_001269_600 |
0.0852511083 |
- |
- |
protein phosphatase 2c, putative [Ricinus communis] |
| 14 |
Hb_001691_200 |
0.0859510673 |
- |
- |
DNA-directed RNA polymerase II subunit RPB2 [Auxenochlorella protothecoides] |
| 15 |
Hb_000409_050 |
0.0864092751 |
transcription factor |
TF Family: HMG |
PREDICTED: FACT complex subunit SSRP1 [Jatropha curcas] |
| 16 |
Hb_011485_060 |
0.0865195412 |
transcription factor |
TF Family: CAMTA |
PREDICTED: calmodulin-binding transcription activator 3 [Jatropha curcas] |
| 17 |
Hb_000896_050 |
0.0873200413 |
- |
- |
PREDICTED: uncharacterized protein LOC105637668 [Jatropha curcas] |
| 18 |
Hb_003159_050 |
0.0873656166 |
- |
- |
PREDICTED: 3-hydroxyisobutyryl-CoA hydrolase-like protein 3, mitochondrial isoform X1 [Jatropha curcas] |
| 19 |
Hb_021576_010 |
0.0874293868 |
- |
- |
PREDICTED: probable protein phosphatase 2C 60 [Jatropha curcas] |
| 20 |
Hb_006210_010 |
0.0875833674 |
- |
- |
PREDICTED: probable acyl-activating enzyme 17, peroxisomal isoform X2 [Jatropha curcas] |