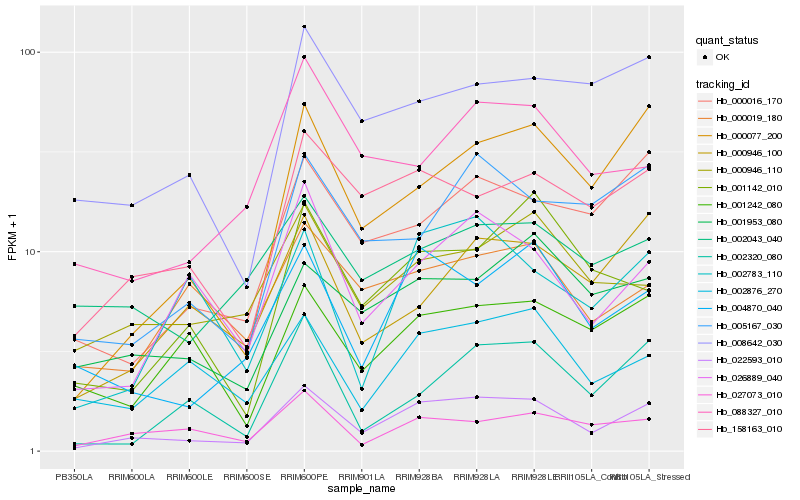
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_022593_010 |
0.0 |
- |
- |
PREDICTED: gamma aminobutyrate transaminase 3, chloroplastic [Jatropha curcas] |
| 2 |
Hb_158163_010 |
0.1504728358 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 3 |
Hb_004870_040 |
0.1505382826 |
- |
- |
- |
| 4 |
Hb_000077_200 |
0.1511442233 |
- |
- |
PREDICTED: thioredoxin [Jatropha curcas] |
| 5 |
Hb_026889_040 |
0.1645339296 |
- |
- |
PREDICTED: heterogeneous nuclear ribonucleoprotein 1 [Jatropha curcas] |
| 6 |
Hb_000016_170 |
0.1650531819 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_001953_080 |
0.1689395814 |
- |
- |
pseudouridine synthase family protein, partial [Manihot esculenta] |
| 8 |
Hb_000946_110 |
0.1725512105 |
- |
- |
GTP cyclohydrolase I, putative [Ricinus communis] |
| 9 |
Hb_005167_030 |
0.1740626935 |
- |
- |
PREDICTED: serine/threonine-protein kinase At5g01020-like, partial [Camelina sativa] |
| 10 |
Hb_002876_270 |
0.1746223004 |
- |
- |
PREDICTED: probable magnesium transporter NIPA4 isoform X1 [Jatropha curcas] |
| 11 |
Hb_008642_030 |
0.176071825 |
- |
- |
PREDICTED: UMP-CMP kinase 3 isoform X1 [Jatropha curcas] |
| 12 |
Hb_000946_100 |
0.17711465 |
- |
- |
PREDICTED: SKI/DACH domain-containing protein 1-like [Jatropha curcas] |
| 13 |
Hb_027073_010 |
0.1784079884 |
- |
- |
PREDICTED: protein CHROMOSOME TRANSMISSION FIDELITY 7 [Jatropha curcas] |
| 14 |
Hb_088327_010 |
0.1816637602 |
- |
- |
PREDICTED: serine hydroxymethyltransferase 7 [Jatropha curcas] |
| 15 |
Hb_002783_110 |
0.1820174947 |
- |
- |
Zeamatin precursor, putative [Ricinus communis] |
| 16 |
Hb_001242_080 |
0.1820181801 |
- |
- |
PREDICTED: UDP-galactose transporter 2-like [Populus euphratica] |
| 17 |
Hb_002320_080 |
0.1820265908 |
- |
- |
PREDICTED: MLO-like protein 13 isoform X2 [Jatropha curcas] |
| 18 |
Hb_002043_040 |
0.1841237182 |
- |
- |
PREDICTED: uncharacterized protein LOC105631509 [Jatropha curcas] |
| 19 |
Hb_000019_180 |
0.1846780088 |
transcription factor |
TF Family: SET |
Histone-lysine N-methyltransferase family protein [Populus trichocarpa] |
| 20 |
Hb_001142_010 |
0.1860550265 |
- |
- |
PREDICTED: alcohol dehydrogenase-like 6 [Jatropha curcas] |