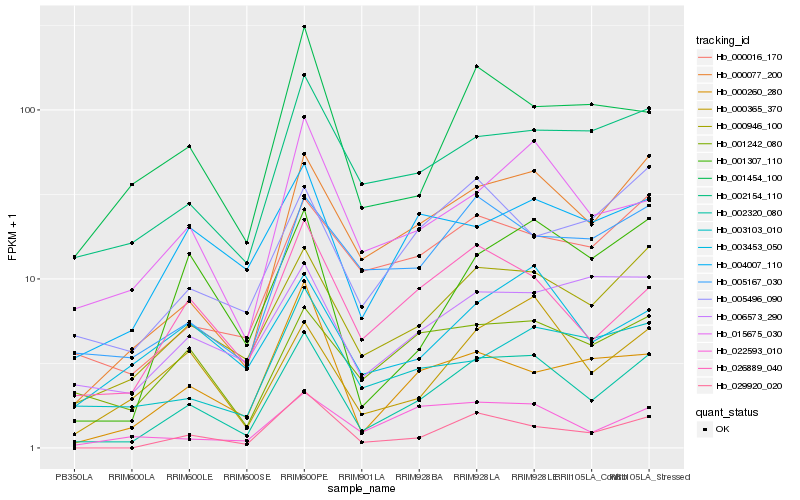
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002320_080 |
0.0 |
- |
- |
PREDICTED: MLO-like protein 13 isoform X2 [Jatropha curcas] |
| 2 |
Hb_000077_200 |
0.1266049112 |
- |
- |
PREDICTED: thioredoxin [Jatropha curcas] |
| 3 |
Hb_000946_100 |
0.1272789392 |
- |
- |
PREDICTED: SKI/DACH domain-containing protein 1-like [Jatropha curcas] |
| 4 |
Hb_029920_020 |
0.139610993 |
- |
- |
PREDICTED: cytokinin riboside 5'-monophosphate phosphoribohydrolase LOG7 [Jatropha curcas] |
| 5 |
Hb_026889_040 |
0.1639336389 |
- |
- |
PREDICTED: heterogeneous nuclear ribonucleoprotein 1 [Jatropha curcas] |
| 6 |
Hb_001307_110 |
0.1669880932 |
- |
- |
PREDICTED: uncharacterized protein LOC105644507 [Jatropha curcas] |
| 7 |
Hb_015675_030 |
0.1696457102 |
- |
- |
PREDICTED: UMP-CMP kinase 3 isoform X2 [Jatropha curcas] |
| 8 |
Hb_000365_370 |
0.1739409646 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_002154_110 |
0.1740705006 |
- |
- |
PREDICTED: protein SPIRAL1-like 1 [Populus euphratica] |
| 10 |
Hb_000016_170 |
0.1768393699 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_005167_030 |
0.178294951 |
- |
- |
PREDICTED: serine/threonine-protein kinase At5g01020-like, partial [Camelina sativa] |
| 12 |
Hb_022593_010 |
0.1820265908 |
- |
- |
PREDICTED: gamma aminobutyrate transaminase 3, chloroplastic [Jatropha curcas] |
| 13 |
Hb_004007_110 |
0.1870119278 |
- |
- |
PREDICTED: mitochondrial outer membrane protein porin 2-like [Jatropha curcas] |
| 14 |
Hb_001242_080 |
0.188399219 |
- |
- |
PREDICTED: UDP-galactose transporter 2-like [Populus euphratica] |
| 15 |
Hb_003103_010 |
0.1886041934 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_005496_090 |
0.1888869435 |
- |
- |
PREDICTED: adenine phosphoribosyltransferase 1-like [Jatropha curcas] |
| 17 |
Hb_001454_100 |
0.1889160057 |
- |
- |
PREDICTED: protein RALF-like 33 [Jatropha curcas] |
| 18 |
Hb_003453_050 |
0.1919628832 |
- |
- |
PREDICTED: calcium-binding mitochondrial carrier protein SCaMC-3-like [Jatropha curcas] |
| 19 |
Hb_006573_290 |
0.192765203 |
- |
- |
PREDICTED: reticulon-like protein B5 [Jatropha curcas] |
| 20 |
Hb_000260_280 |
0.1930222672 |
- |
- |
auxin:hydrogen symporter, putative [Ricinus communis] |