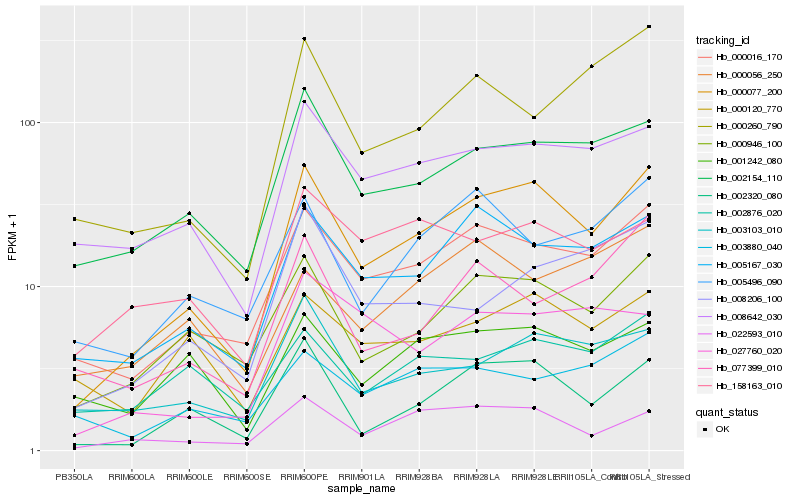
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000077_200 |
0.0 |
- |
- |
PREDICTED: thioredoxin [Jatropha curcas] |
| 2 |
Hb_000016_170 |
0.1159168951 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_002320_080 |
0.1266049112 |
- |
- |
PREDICTED: MLO-like protein 13 isoform X2 [Jatropha curcas] |
| 4 |
Hb_000946_100 |
0.1340635913 |
- |
- |
PREDICTED: SKI/DACH domain-containing protein 1-like [Jatropha curcas] |
| 5 |
Hb_005167_030 |
0.1343832914 |
- |
- |
PREDICTED: serine/threonine-protein kinase At5g01020-like, partial [Camelina sativa] |
| 6 |
Hb_002154_110 |
0.1450453848 |
- |
- |
PREDICTED: protein SPIRAL1-like 1 [Populus euphratica] |
| 7 |
Hb_022593_010 |
0.1511442233 |
- |
- |
PREDICTED: gamma aminobutyrate transaminase 3, chloroplastic [Jatropha curcas] |
| 8 |
Hb_008642_030 |
0.1512236312 |
- |
- |
PREDICTED: UMP-CMP kinase 3 isoform X1 [Jatropha curcas] |
| 9 |
Hb_003103_010 |
0.1629056285 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_005496_090 |
0.17080626 |
- |
- |
PREDICTED: adenine phosphoribosyltransferase 1-like [Jatropha curcas] |
| 11 |
Hb_000260_790 |
0.1735460768 |
- |
- |
NADH dehydrogenase 1 alpha subcomplex subunit 12 isoform 1 [Theobroma cacao] |
| 12 |
Hb_008206_100 |
0.1749247522 |
- |
- |
PREDICTED: uncharacterized protein LOC105638302 [Jatropha curcas] |
| 13 |
Hb_158163_010 |
0.1759986034 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 14 |
Hb_000120_770 |
0.1778010056 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_077399_010 |
0.1797857413 |
- |
- |
hypothetical protein POPTR_0003s09910g [Populus trichocarpa] |
| 16 |
Hb_001242_080 |
0.1821089538 |
- |
- |
PREDICTED: UDP-galactose transporter 2-like [Populus euphratica] |
| 17 |
Hb_027760_020 |
0.1839055836 |
- |
- |
hypothetical protein M569_04462, partial [Genlisea aurea] |
| 18 |
Hb_002876_020 |
0.185439935 |
- |
- |
PREDICTED: maf-like protein DDB_G0281937 isoform X2 [Jatropha curcas] |
| 19 |
Hb_003880_040 |
0.1854707103 |
- |
- |
hypothetical protein POPTR_0001s30770g [Populus trichocarpa] |
| 20 |
Hb_000056_250 |
0.1913927118 |
- |
- |
PREDICTED: haloacid dehalogenase-like hydrolase domain-containing protein 3 [Jatropha curcas] |