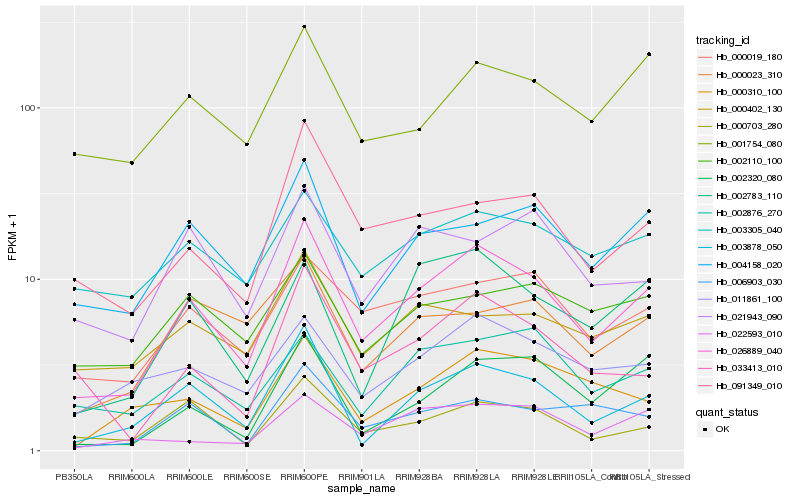
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_026889_040 |
0.0 |
- |
- |
PREDICTED: heterogeneous nuclear ribonucleoprotein 1 [Jatropha curcas] |
| 2 |
Hb_003878_050 |
0.1338324163 |
- |
- |
PREDICTED: peptide-N4-(N-acetyl-beta-glucosaminyl)asparagine amidase A-like [Jatropha curcas] |
| 3 |
Hb_000019_180 |
0.1365528648 |
transcription factor |
TF Family: SET |
Histone-lysine N-methyltransferase family protein [Populus trichocarpa] |
| 4 |
Hb_004158_020 |
0.1399193381 |
- |
- |
Rab1 [Hevea brasiliensis] |
| 5 |
Hb_091349_010 |
0.1444825912 |
- |
- |
hypothetical protein EUGRSUZ_C02287 [Eucalyptus grandis] |
| 6 |
Hb_000703_280 |
0.1448950406 |
- |
- |
o-linked n-acetylglucosamine transferase, ogt, putative [Ricinus communis] |
| 7 |
Hb_011861_100 |
0.1478181437 |
- |
- |
amino acid transporter, putative [Ricinus communis] |
| 8 |
Hb_002783_110 |
0.1505967956 |
- |
- |
Zeamatin precursor, putative [Ricinus communis] |
| 9 |
Hb_033413_010 |
0.1512087556 |
- |
- |
Reticuline oxidase precursor, putative [Ricinus communis] |
| 10 |
Hb_006903_030 |
0.1534051509 |
- |
- |
PREDICTED: uncharacterized protein LOC105640666 [Jatropha curcas] |
| 11 |
Hb_001754_080 |
0.1583146126 |
- |
- |
PREDICTED: protein RALF-like 33 [Jatropha curcas] |
| 12 |
Hb_002876_270 |
0.1586227876 |
- |
- |
PREDICTED: probable magnesium transporter NIPA4 isoform X1 [Jatropha curcas] |
| 13 |
Hb_000402_130 |
0.1606363055 |
- |
- |
PREDICTED: AP-1 complex subunit mu-2 [Jatropha curcas] |
| 14 |
Hb_003305_040 |
0.1618833422 |
- |
- |
AP47/50p mRNA family protein [Populus trichocarpa] |
| 15 |
Hb_021943_090 |
0.1629534309 |
- |
- |
PREDICTED: probable methylenetetrahydrofolate reductase [Jatropha curcas] |
| 16 |
Hb_000023_310 |
0.1629831287 |
- |
- |
PREDICTED: probable folate-biopterin transporter 2 [Populus euphratica] |
| 17 |
Hb_002320_080 |
0.1639336389 |
- |
- |
PREDICTED: MLO-like protein 13 isoform X2 [Jatropha curcas] |
| 18 |
Hb_002110_100 |
0.1639543521 |
- |
- |
PREDICTED: pyridoxine/pyridoxamine 5'-phosphate oxidase 1, chloroplastic [Jatropha curcas] |
| 19 |
Hb_000310_100 |
0.1643776318 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_022593_010 |
0.1645339296 |
- |
- |
PREDICTED: gamma aminobutyrate transaminase 3, chloroplastic [Jatropha curcas] |